DML Analysis Part 27
Resolving DML and DMR visualization issues
TL;DR Certain things are a dumpster fire so it’s time to put out the dumpster fire.
Quick overview
I looked at two different chromosomes to see what was happening. Here are some things I learned:
- The gene background is hot flaming garbage.


It should just be CG motifs that have 3x coverage or more, but instead it includes multiple loci that aren’t cytosines.
- The DML overlap CG motifs better.


I mildly trust the DML track.
- The 100 bp DMR are inconsistent
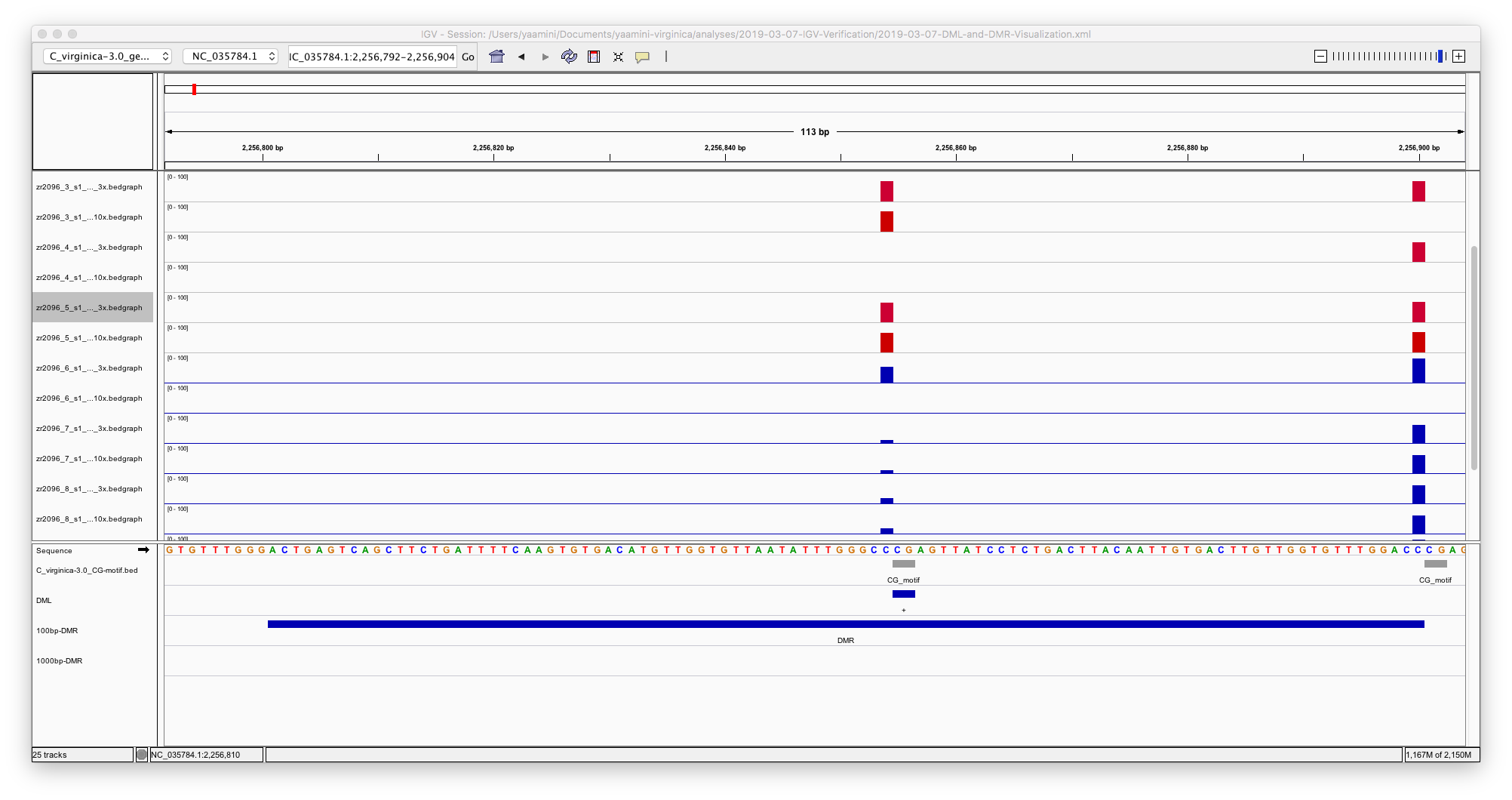

Sometimes DMR include mulitple CG motifs or DML, but sometimes they don’t. I think using a step size of 100 bp may be influencing this (more on that below).
- The 1000 bp DMR make no sense


The 1000 bp DMR are heavily influenced by regions where one sample is hypermethylated in that region, but other samples aren’t methylated there (or have no data). I did find an instance or two where multiple samples were hypermethylated in a 1000 bp DMR, but these were rare over the two chromosomes I looked at.
Generate 5x and 10x DML tracks
At this point, I mildly trust DML. We’re currently using 3x coverage for analyses, but previous papers have used 5x coverage. We decided to look at 5x and 10x DML tracks as well.
In this R Markdown file, I created 5x and 10x DML bedfiles (found here). Steven pointed out that he used destrand = TRUE in his unite command, and I did not. My current stranded output includes + or - indications for forward and reverse strands, but normal discussion of methylated loci does not include strandeness. According to the methylKit manual, destrand = TRUE provides better coverage for CpG methylation. I created 5x and 10x coverage tracks using both destrand = FALSE, the default, and destrand = TRUE. To visualize everything in my IGV session, I also created 5x sample coverage tracks, found here.
- Some DML were retained through all coverage types.

I didn’t see much loss going from 3x to 5x DML tracks, but there were more DML lost from 5x to 10x.
- Some DML were ony present in destranded 5x or 10x tracks.

This is probably a result of the increased coverage in the destranded tracks.
- I also found something really weird.

This CpG had a DML on the forward strand in the 3x, 5x, and destranded tracks, but on the reverse strand in the 10x track. Not sure how this can happen…
Going forward
- Figure out which DML track to use for remaining analyses and characterize DML locations in that track
- Describe methylation irrespective of treatment
- Work through gene-level analysis
- Figure out what’s going on with DMR
- Figure out what’s going on with the gene background