DML Analysis Part 14
Tiling window analysis in methylKit
Now that I’m using mincov = 3, I wanted to try the tiling window analysis in methylKit. I added code for the analysis to the end of this R Markdown file.
A tiling window analysis will allow me to identify differentially methylated regions to complement the differentially methylated loci I previously identified. The analysis takes a given window (default is 1000 bp) and adds the number of C and T from each covered cytosine, and spits out a total number of C and T for each tile. I then takes a step (default is 1000 bp) and peforms the same analysis on the next tile.
I chose to do this analysis with three different window-step size combinations:
- 100 bp window and step size
- 1000 bp window and step size (the
methylKitdefault) - 1000 bp window and 100 bp step size
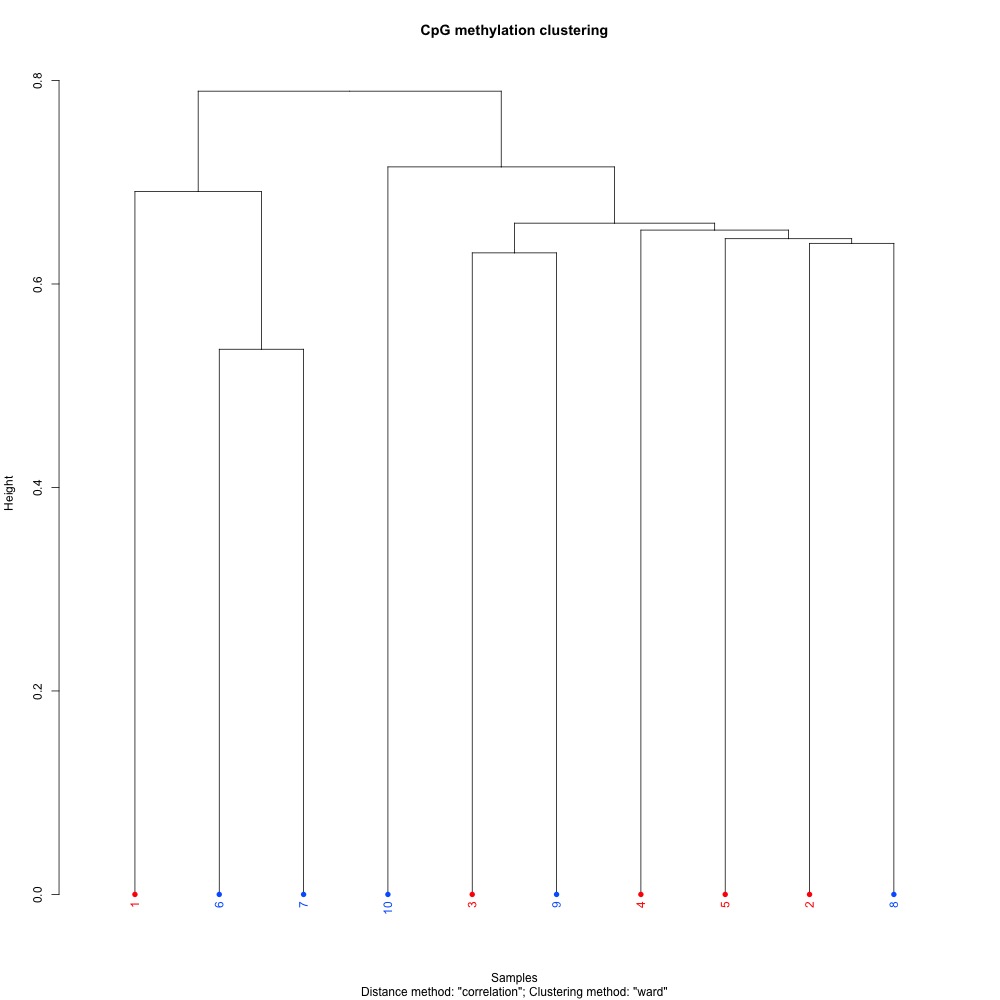
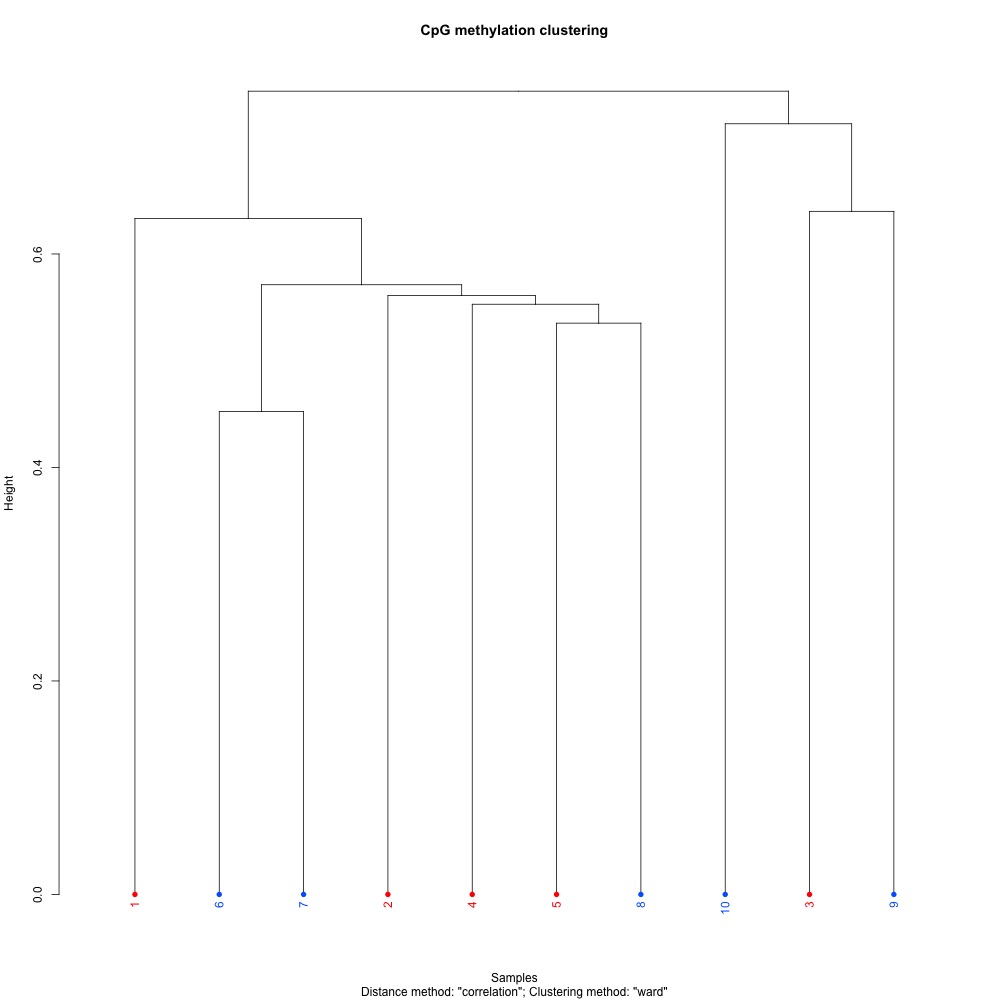
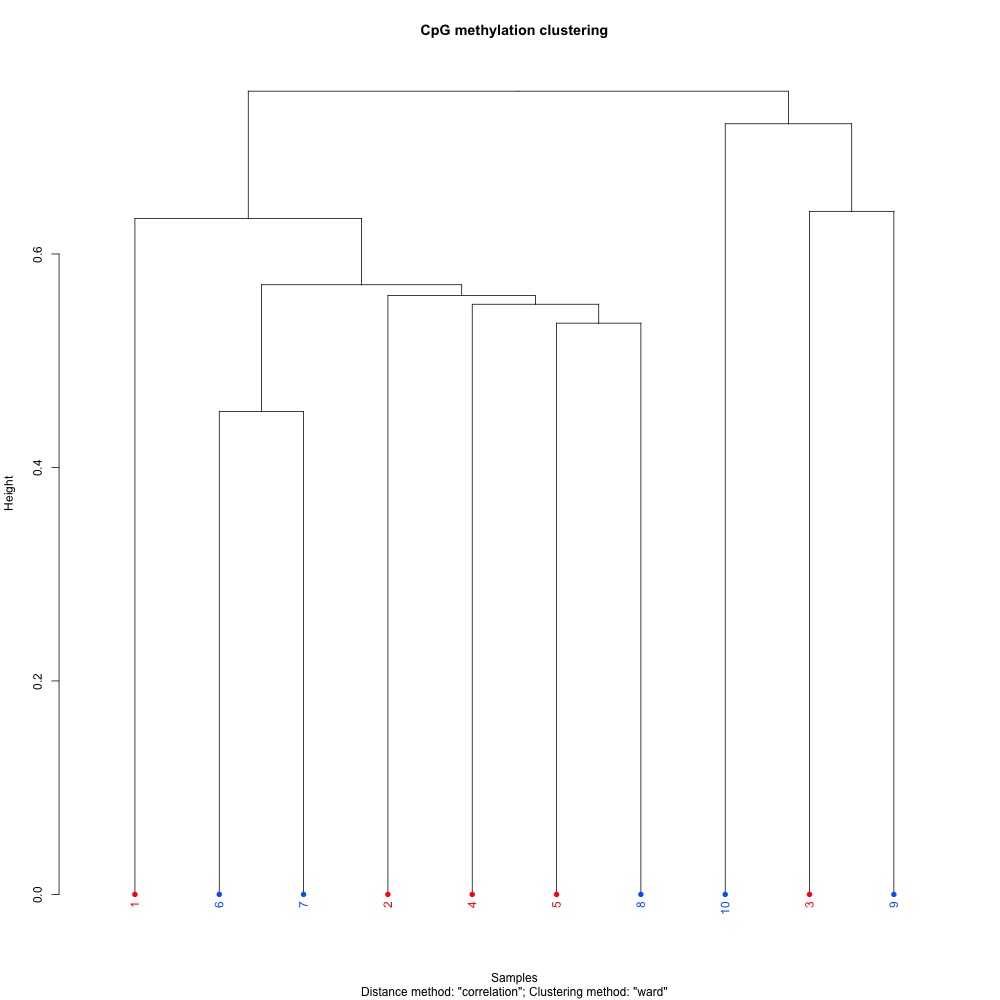
Figures 1-3. Full sample CpG methylation clustering for 1) 100 bp window and step size 2) 1000 bp window and step size and 3) 1000 bp window and 100 bp step size.
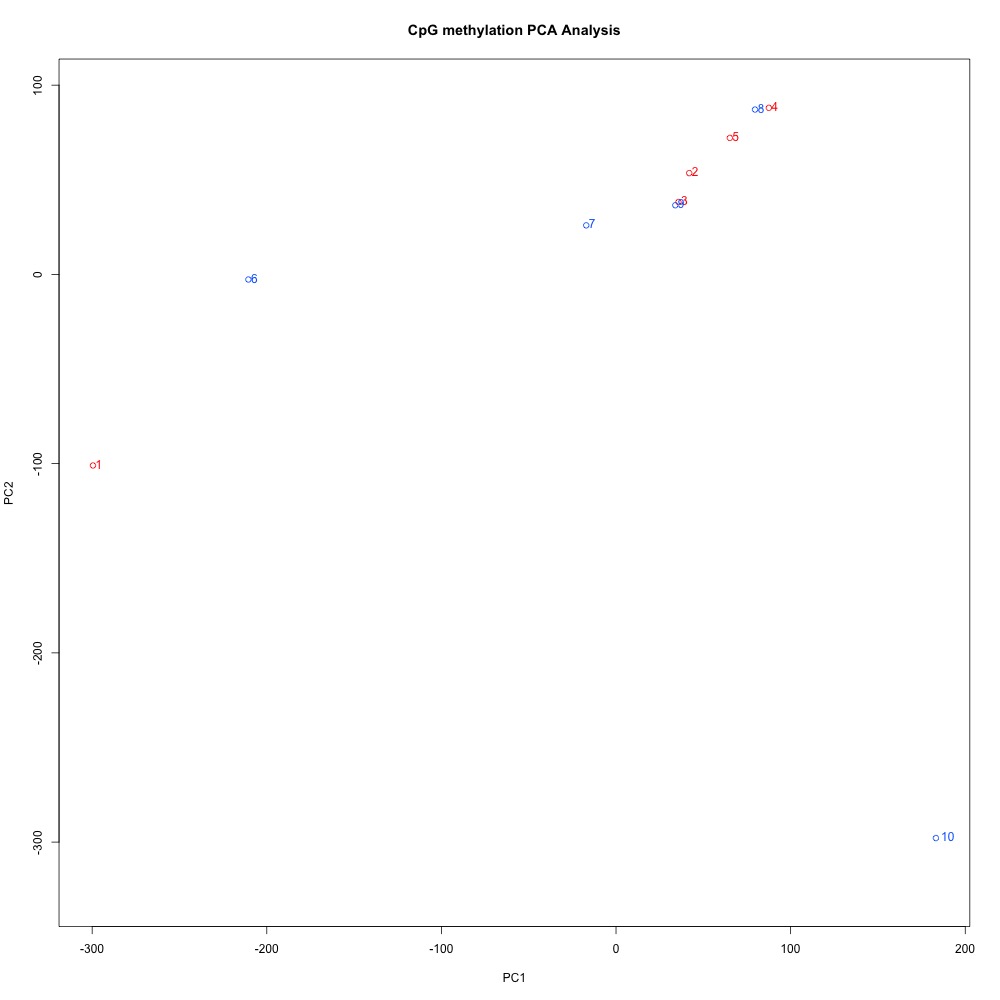
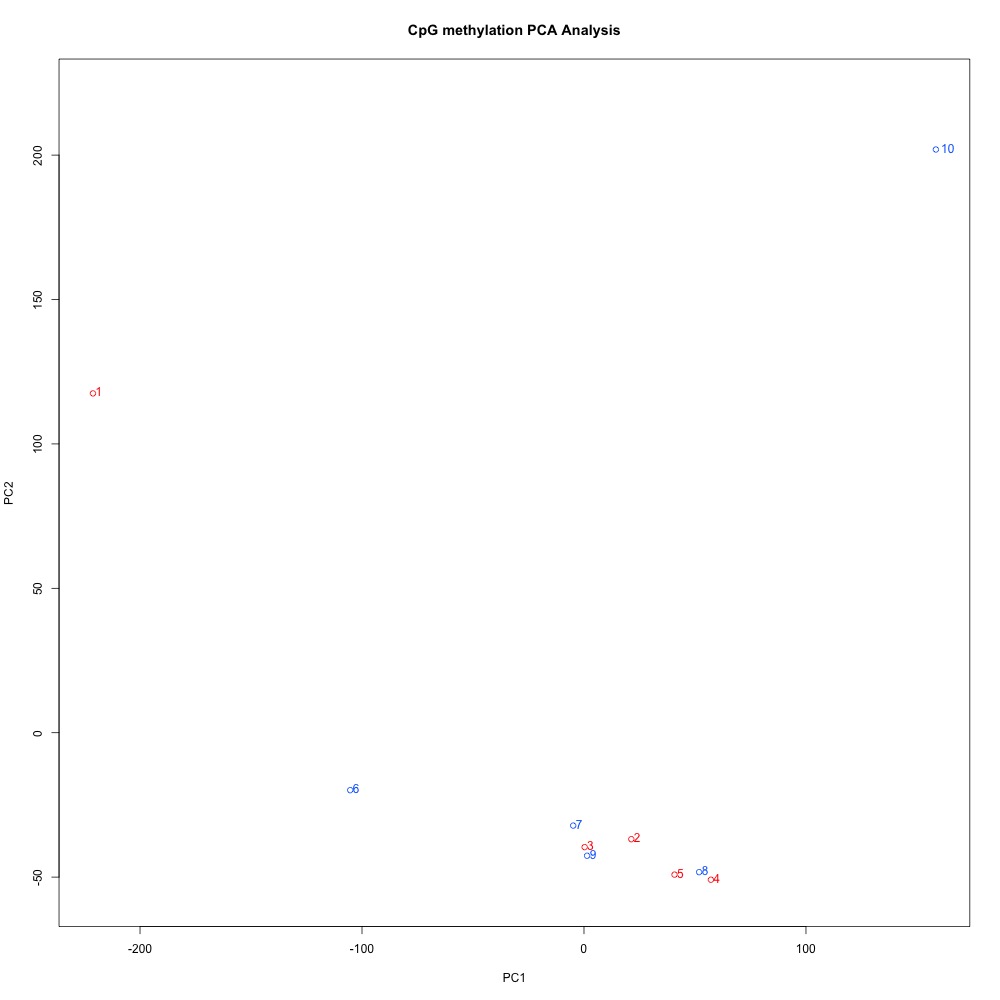
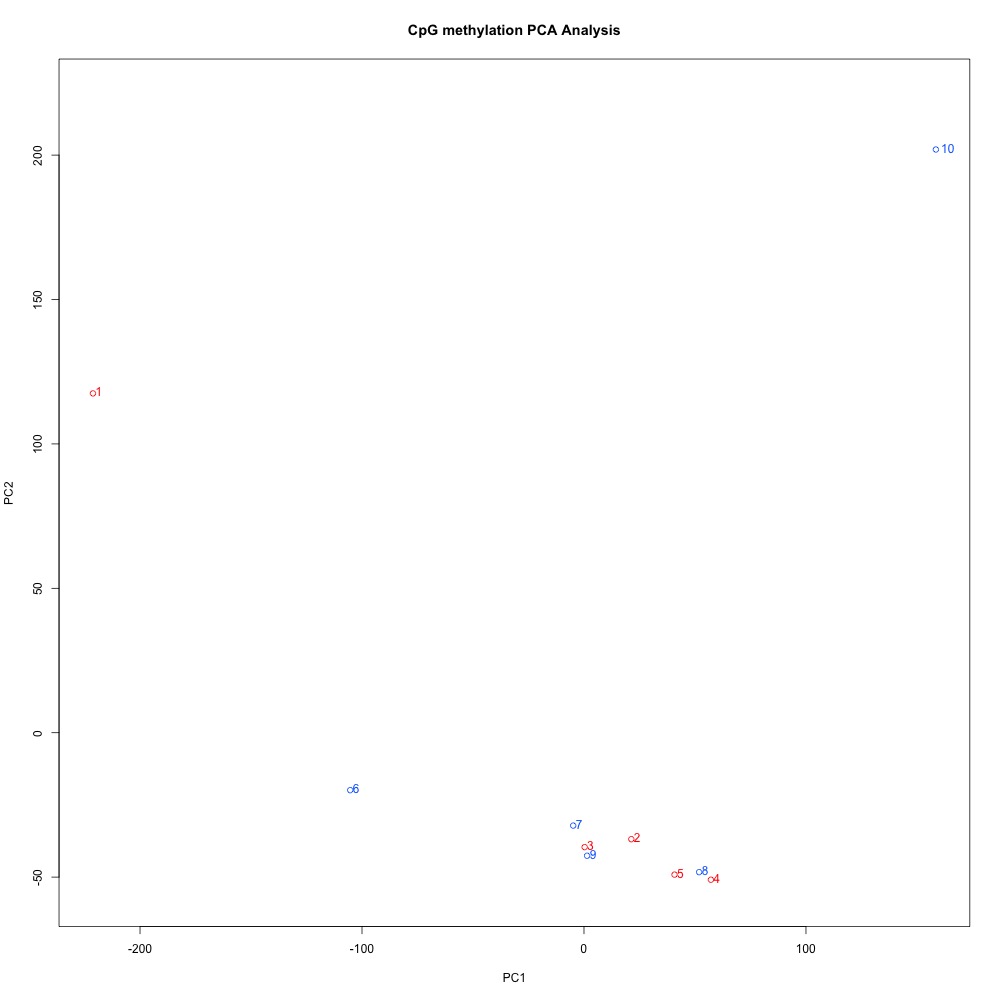
Figures 4-6. PCA of full sample methylation for 1) 100 bp window and step size 2) 1000 bp window and step size and 3) 1000 bp window and 100 bp step size.
These clustering plots are slightly different from same kind associated with DMLs:
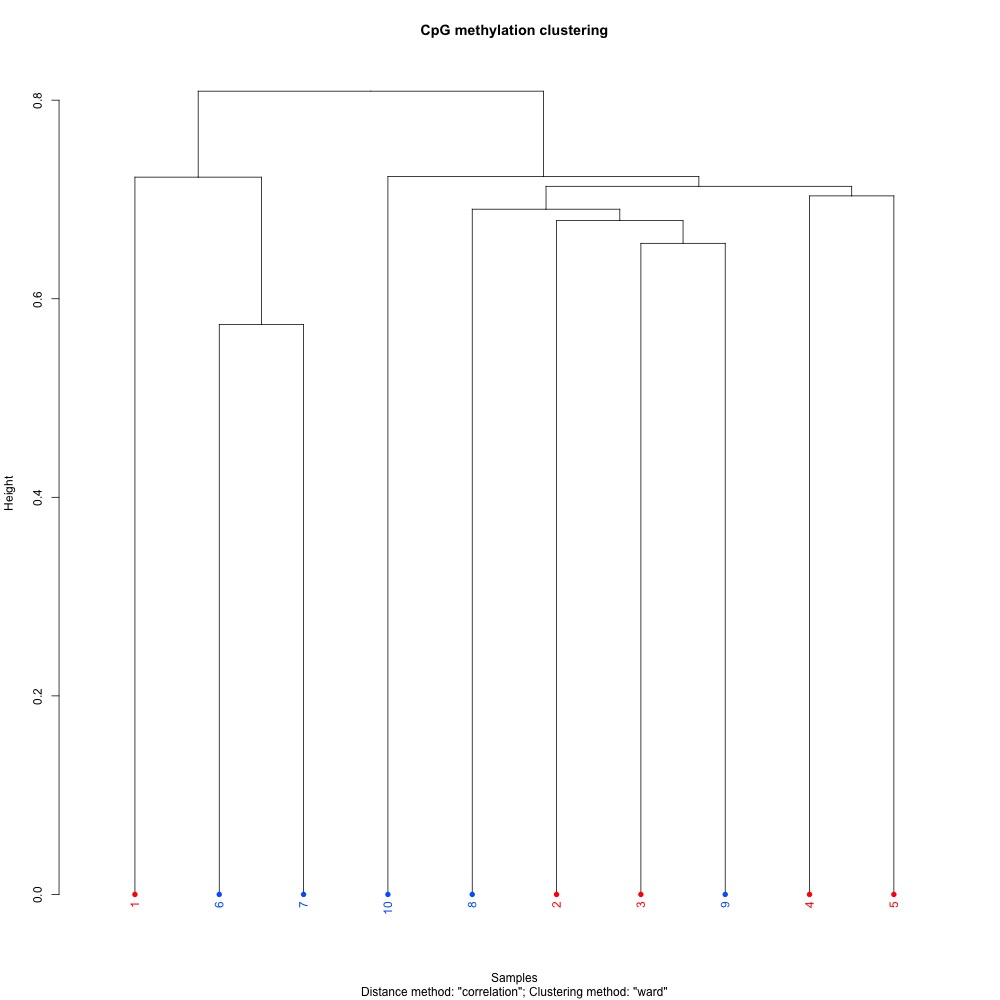
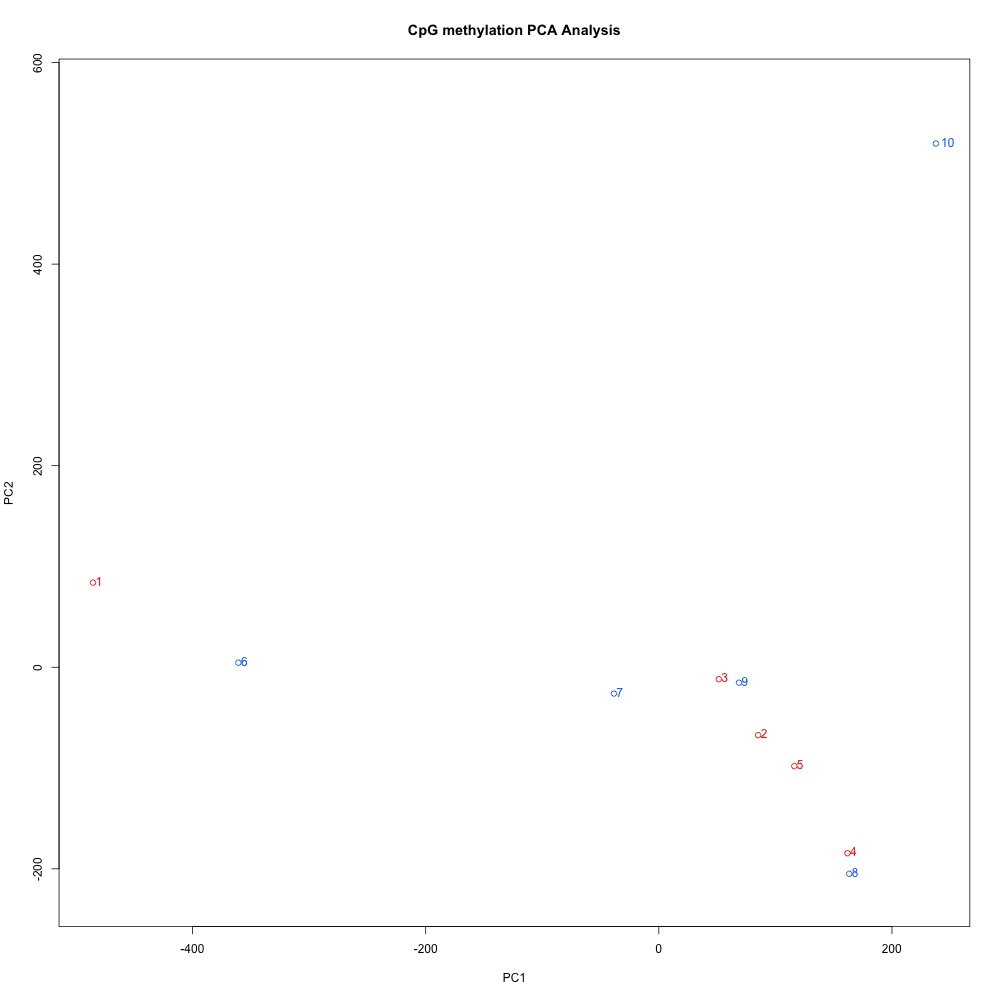
Figures 7-8. Full sample CpG methylation clustering and PCA of full sample methylation using mincov = 3.
When calcuating differential methylation, I didn’t get any error about a glm not fitting. I did get an error when I was calculating differential methylation for individual loci.
Table 1. Window size, step size, total number of regions produced, and the number of DMLs that were at least 50% different between treatment and control samples. The number of regions and siginificantly different DMRs seem to be dictated by the window size, and not the step size.
| Window Size (bp) | Step Size (bp) | Total Regions | Number of Significantly Different DMRs |
|---|---|---|---|
| 100 | 100 | 217538 | 162 |
| 1000 | 1000 | 104144 | 118 |
| 1000 | 100 | 104144 | 118 |
I wrote out DMRs for each method in individual .csv files:
Going forward
I think it makes sense to have smaller-sized DMRs, so I’d rather continue wiht the 100 bp windows and steps. The 100 bp window and step PCA also has the best separation between ambient and treatment samples. I posted this issue to get Steven’s opinion, and he agreed. Now to get a list of genes!