Correlating Technical Replicates Part 5
Things DO get better! (Kind of)
Steven took a reformatted dataset I made with this script yesterday and calculated the coefficient of variance between each technical replicate for all my transitions. He then eliminated all transitions with a CV greater than 20 and saved the document. Today, I averaged my technical replicates and proceeded with my NMDS and ANOSIM analyses to see if anything had changed in this script.
I found that there was still no clear separation of proteins, but Skokomish River seemed to be a bit more separated from the general large cluster. My ANOSIM results were not significant, so I know statistically there is no separation occuring by site or eelgrass condition.
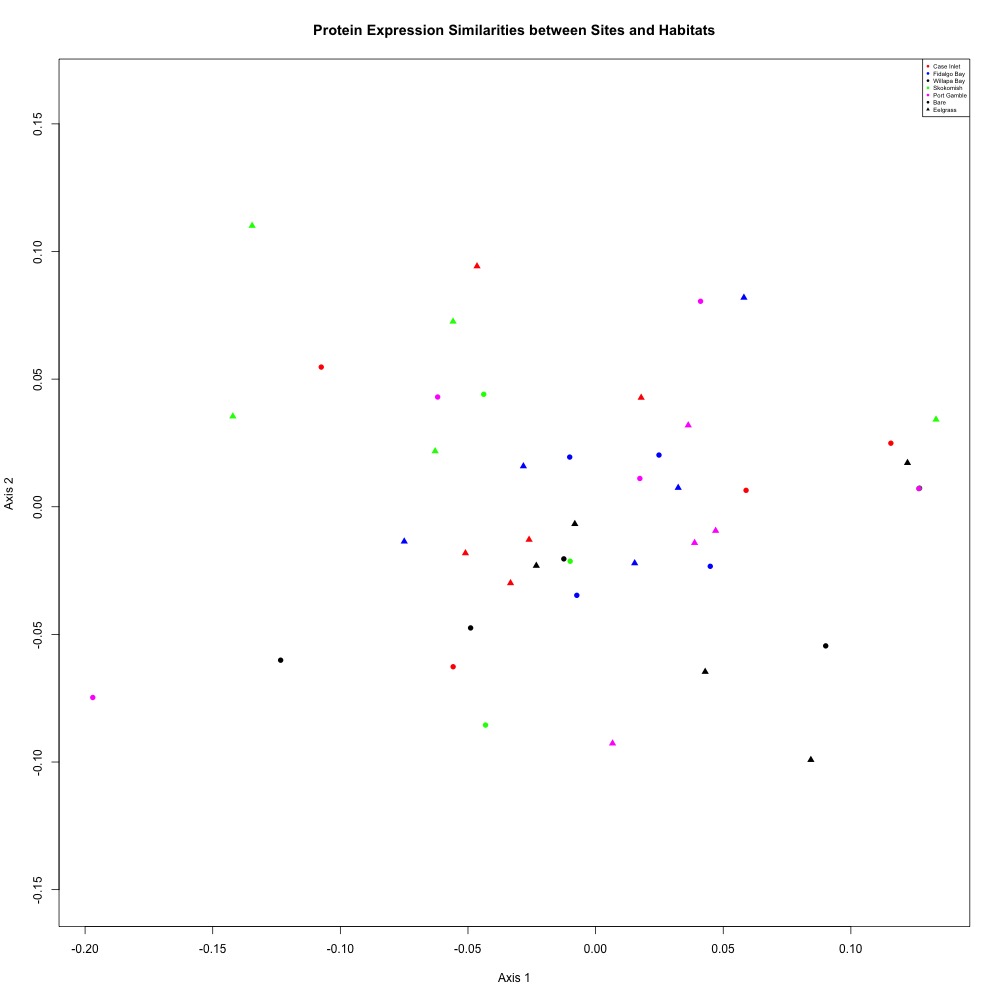
Figure 1. NMDS for averaged technical replicates after CV filtering.
I then decided to look more closely at the dataset that Steven created and see how it affected the clustering of my technical replicates in this script. This clustering is much better, but still not as good as it needs to be. While my samples are clustering together, they’re supposed to be right on top of eachother.
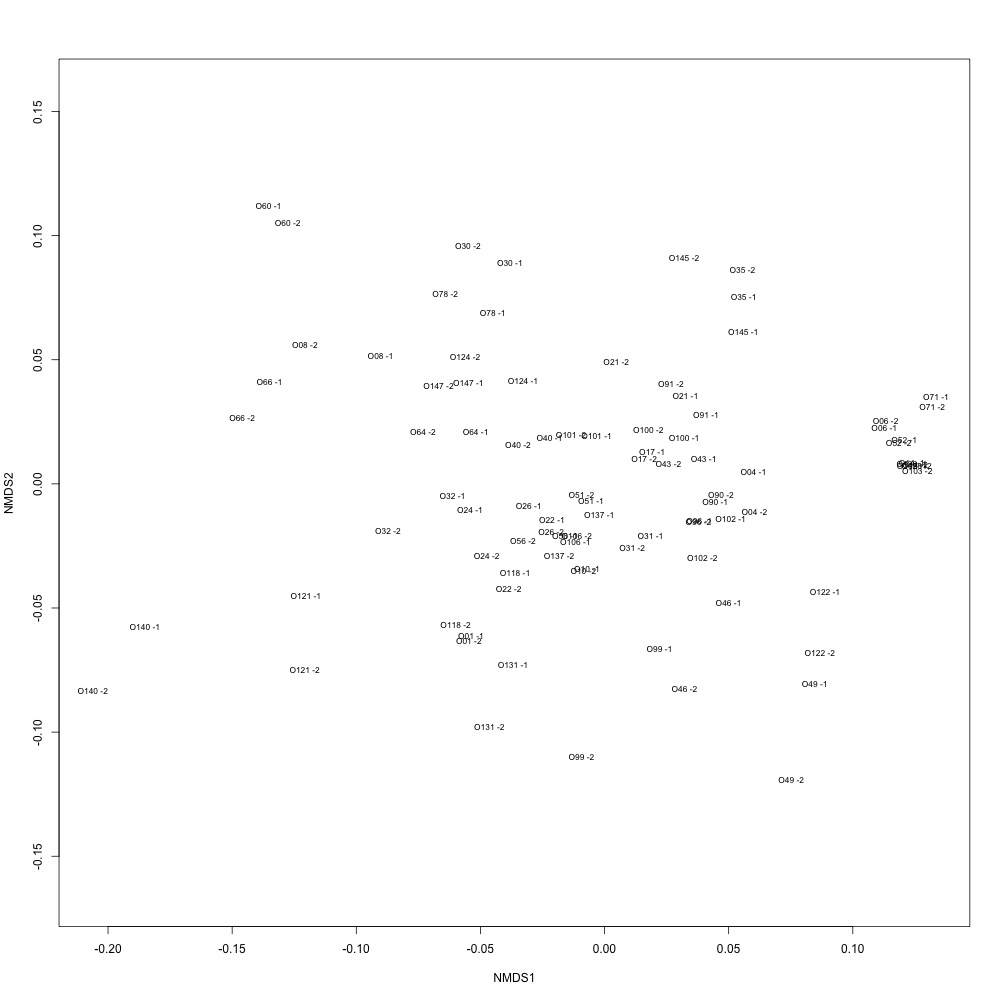
Figure 2. NMDS for technical replication.
I calculated the ordination distances between technical replicates as well.
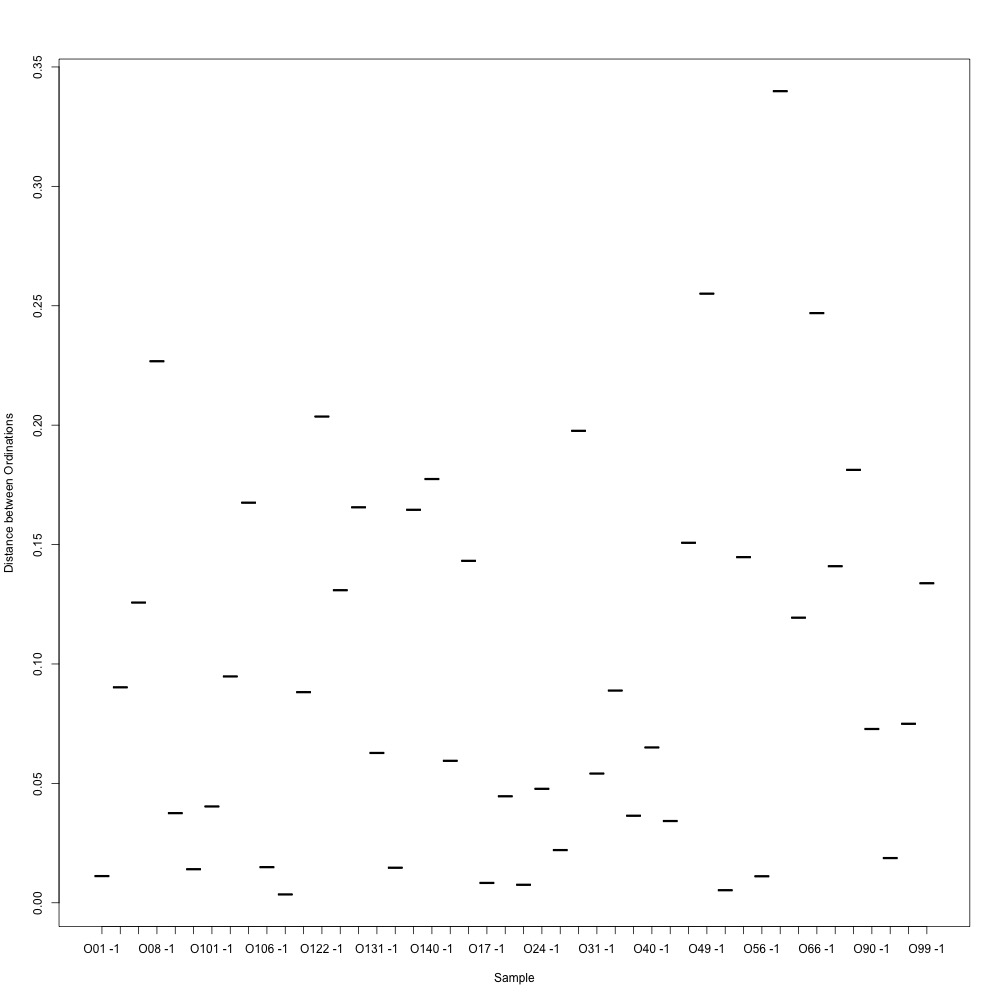
Figure 3. Ordination distances been technical replicates.
I decided to use a cutoff of 0.2 to remove the 10% of my samples with the highest ordination distances. After removing these samples from analysis, I averaged my technical replicates and remade my NMDS plot. Once again, there wasn’t any clear separation. The Skokomish River points I thought might be separating out turned out to be some of the outliers that I pulled. Welp.
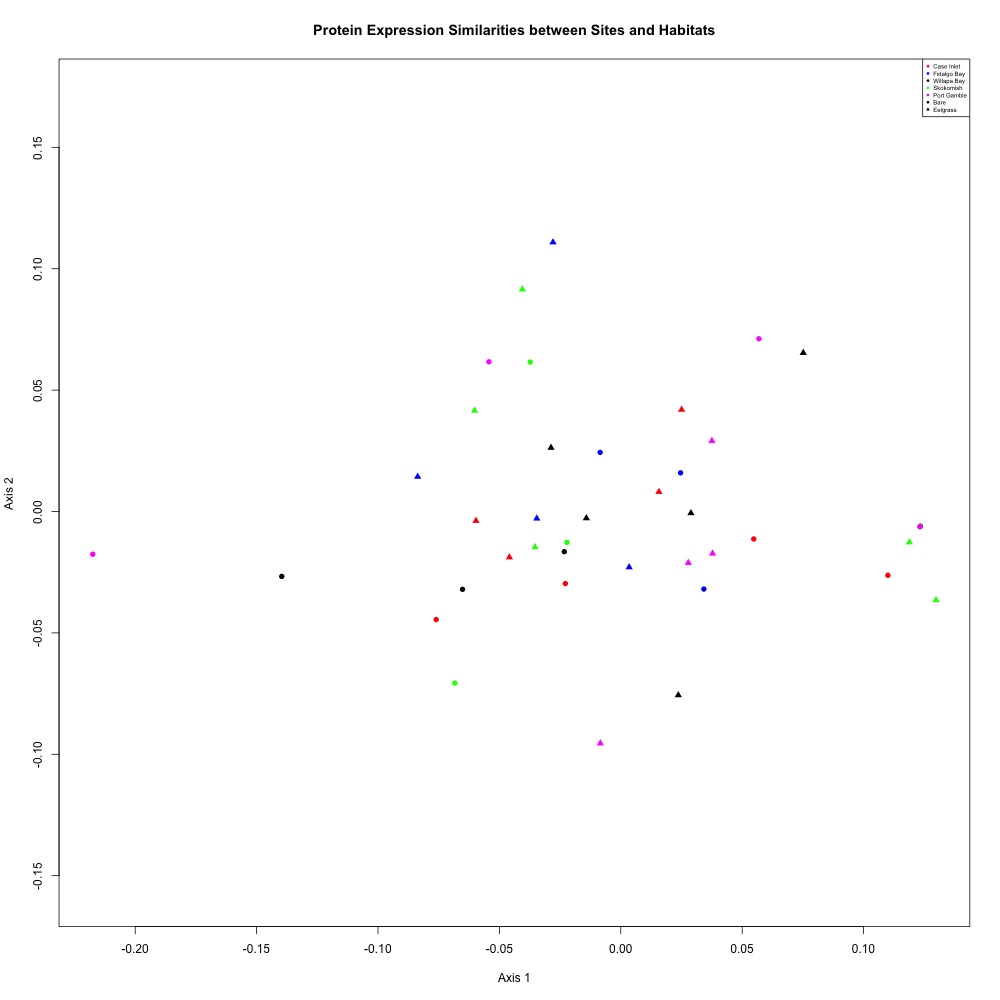
Figure 4. NMDS for averaged technical replicates after CV and ordination distance filtering.
This process demonstrated that using coefficients of variance can help improve my technical replication. I didn’t look too closely at Steven’s list, so I don’t know if there were some proteins with only one transition after filtering that should be removed too. I wonder if using a stricter cutoff will improve things even more? I’m going to look at my regression cutoffs and reaverage my technical replicates to see if other methods change my outcomes. After giving everything a cursory look, I can figure out how to proceed.