Correlating Technical Replicates Part 6
Dead ends and new plans
Dead end:
I went through most of my process to understand if an R-squared cutoff could help improve the statistical accuracy of my technical replication. I was able to average my technical replicates and make an NMDS p lot and ANOSIM (not significant) in this script.
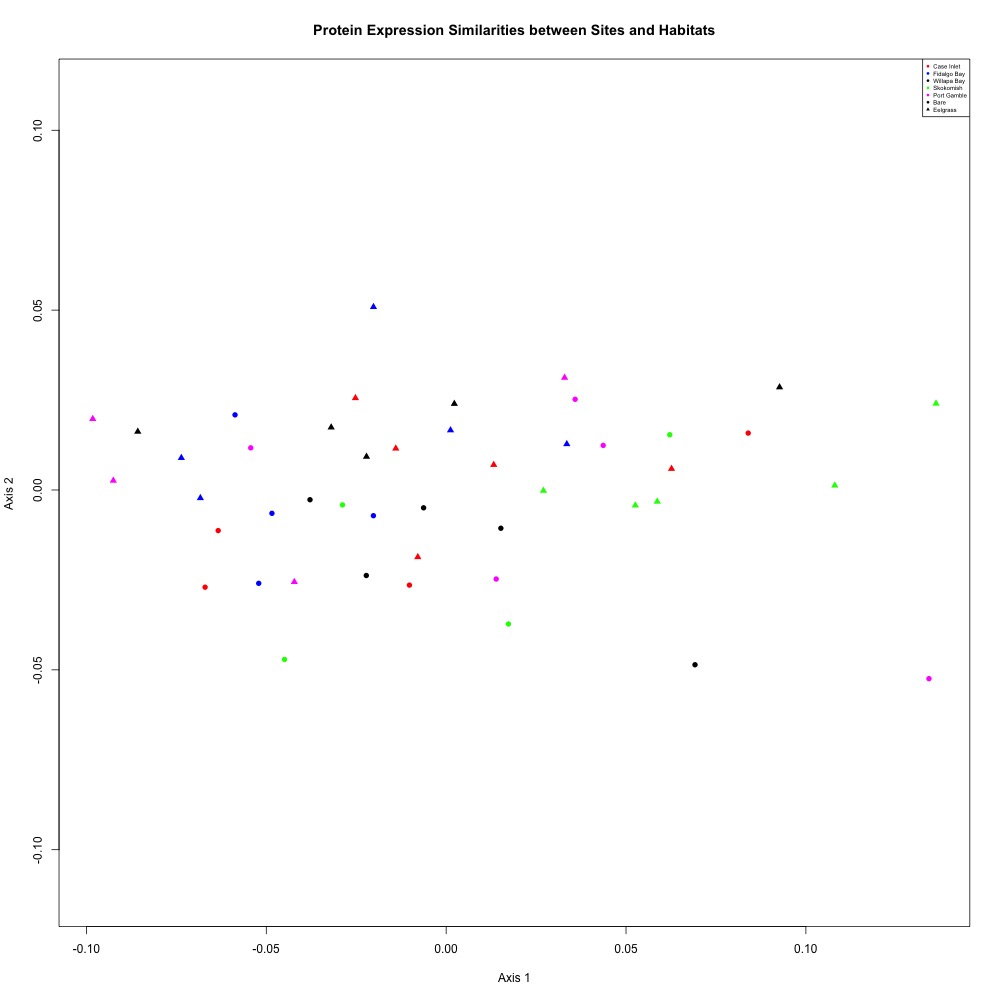
Figure 1. Nonsignificant NMDS after filtering transitions with an R-squared coefficient less than 0.6
I was going to try and remove samples with large ordination distances, but Steven pointed out that an R-squared value can meaningless when it comes to technical replication quality.
New plan:
I’m going to filter my transitions using coefficient of variance cutoffs of 15 and 10. I’ll then see how these impact my NMDS and ANOSIM analyses. I’ll also make boxplots for each transition I retain like I did for PCSGA. This will allow us to understand how expression is varying at a site level. I’m not conviced I’ll have any differences based on site or eelgrass condition, but even a null result is a good result when it’s statistically sound!