DML Analysis Part 8
DML-mRNA Gene Enrichment Results
Yesterday, I went through DML-mRNA overlap gene enrichment in DAVID (see my lab notebook post), but I was unable to generate plots in ReVIGO. Turns out my issue with ReVIGO was related to Adobe Flash. Once I updated my flash player, I was able to create the interactive graphics. I added the graphics and explanations below to my R Markdown file as well. I probably need to learn how to use the R script to generate plots for reproducibility purposes, but that can wait.
I pasted significant GOterms and p-values into ReViGO to generate the following figures. For each category, I created a plot with medium term similarity, and either sizes based on logSize or log10p-value.
Biological processes:
Processes include cytoskeleton and microtubule movement, cell cycle, and protein activity. The most interesting results were those related to DNA and reproduction. DNA replication, mRNA processing, positive regulation of transcription from RNA polymerase II promoters, and response to DNA damage were all enriched. Additionally, enrichment positive regulation of histone H3-K9 acetylation, methylation-dependent chromatin silencing, DNA methylation involved in gamete generation, and fertilization point to mechanisms for epigenetic inheritance! This is going to be the central finding of my PCSGA talk.
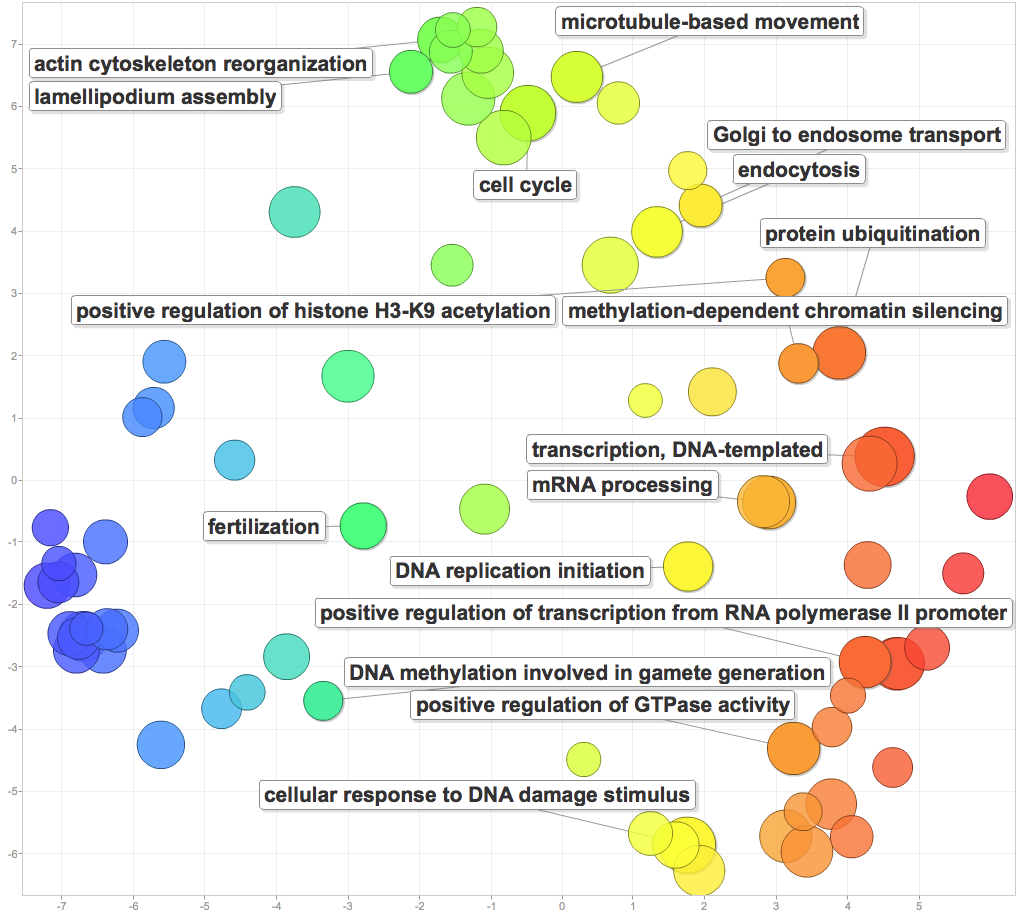
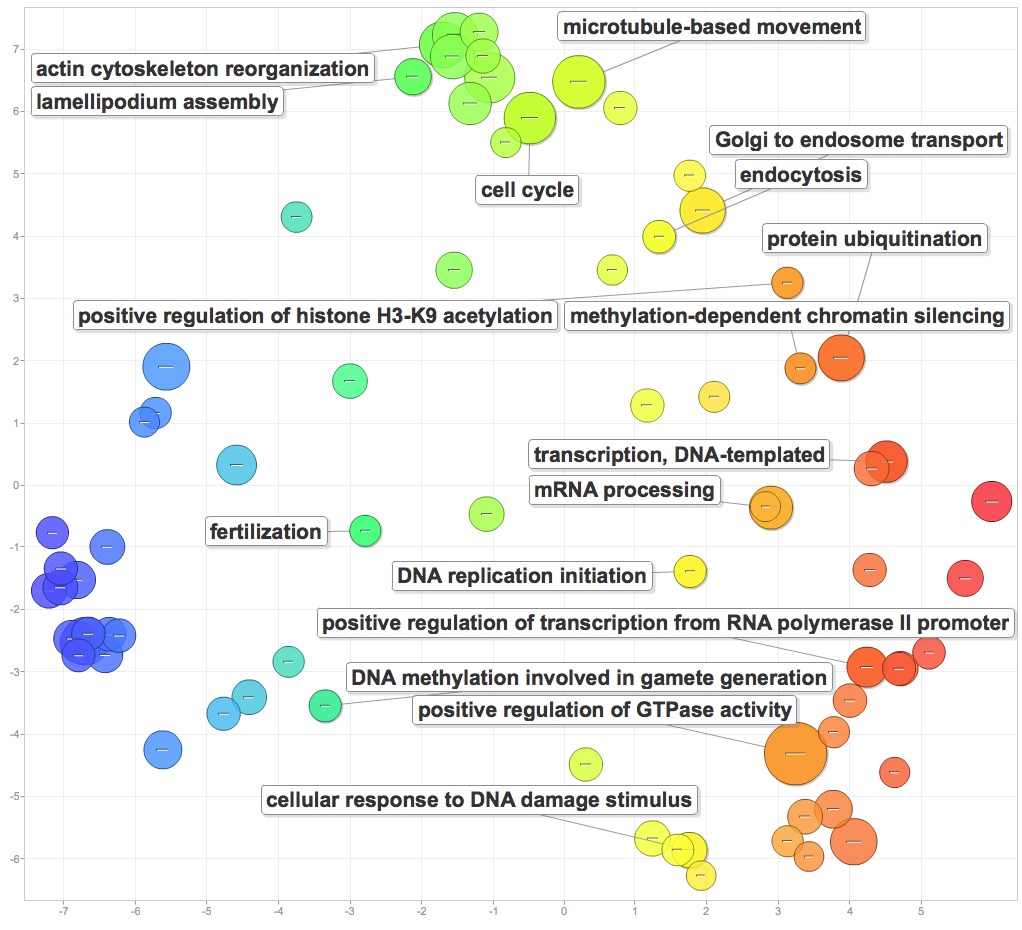
Cellular components:
Cellular components involved in enriched processes included protein complexes, transcription factor complexes, chromosomes, centromeres, microtubules, and cilia.
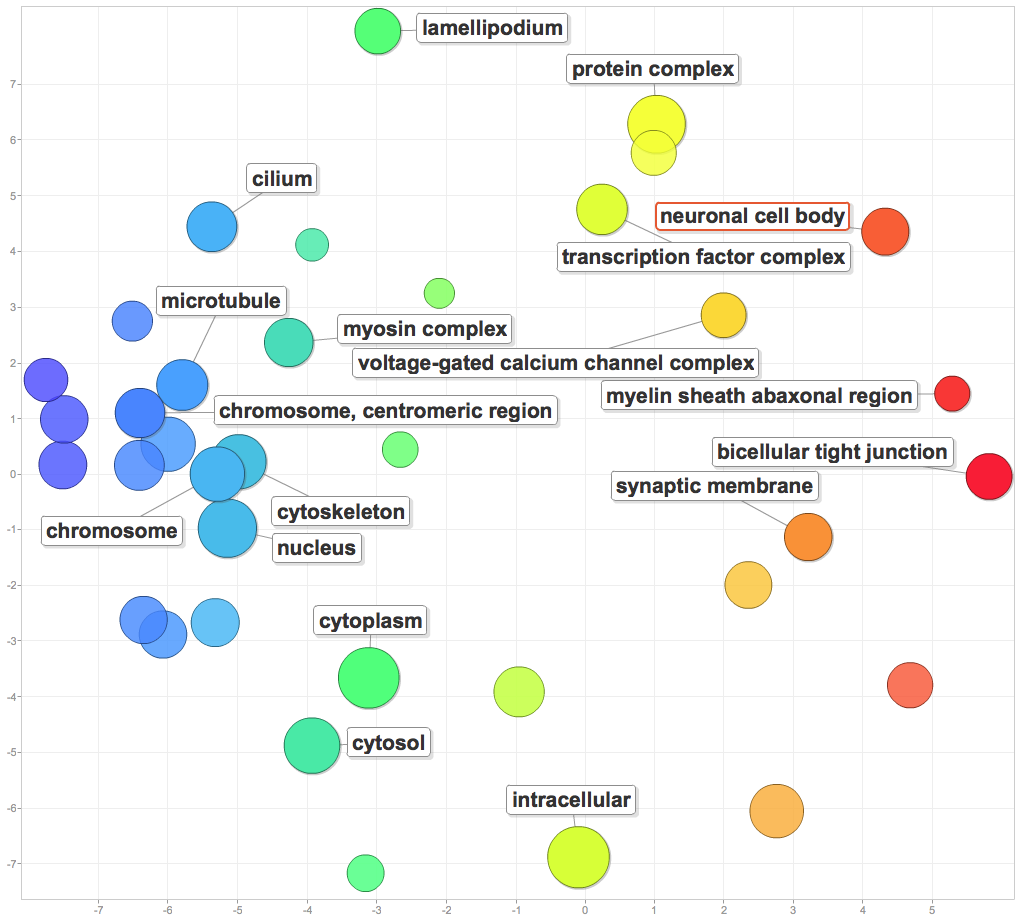
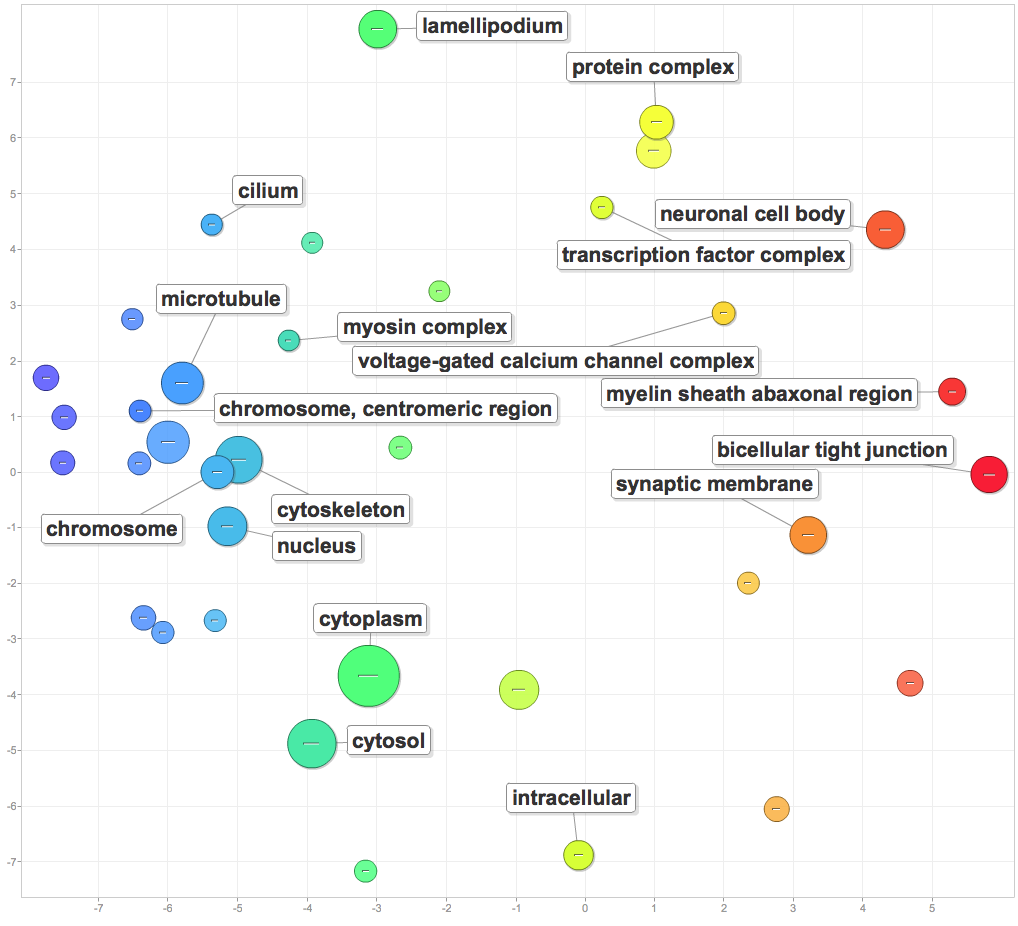
Molecular function:
Molecular functions of enriched processes include GTPase and ATPase activity, helicase activity, microtuble motor activity, protein kinase binding and activity, transcription regulatory region DNA binding, general DNA binding, and ion binding.
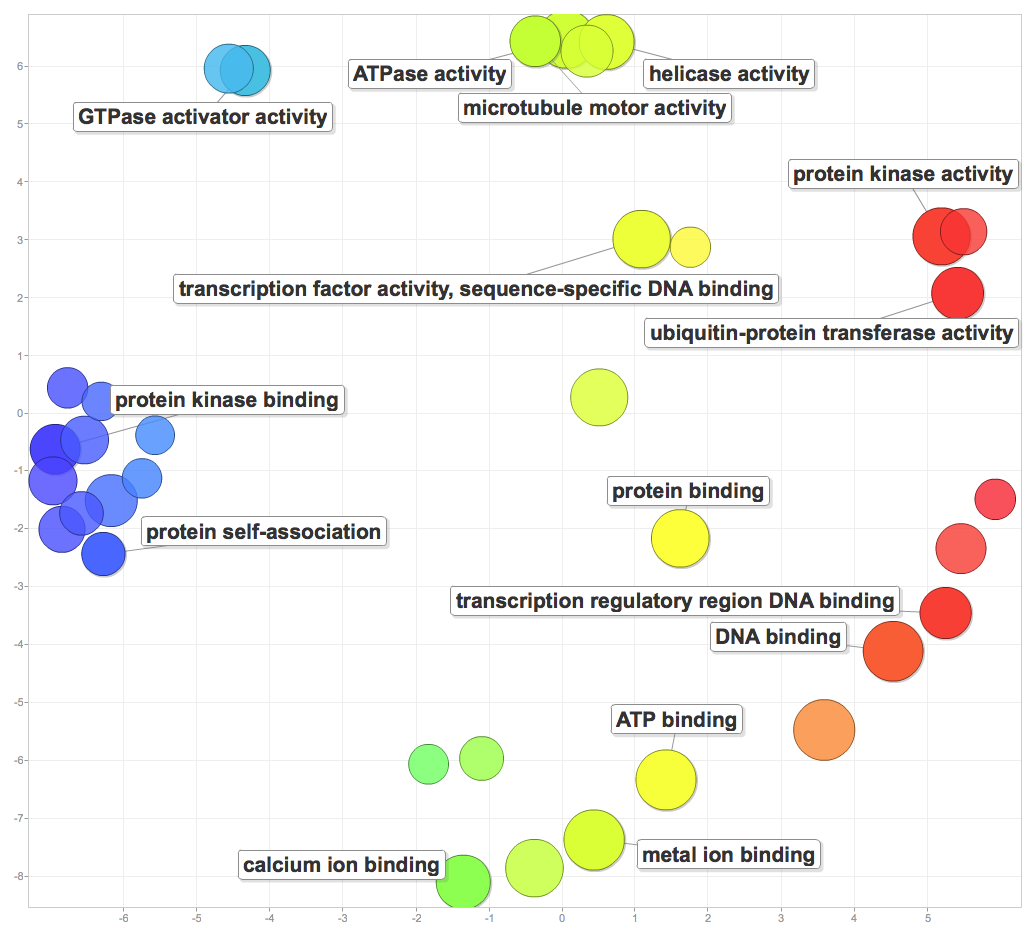
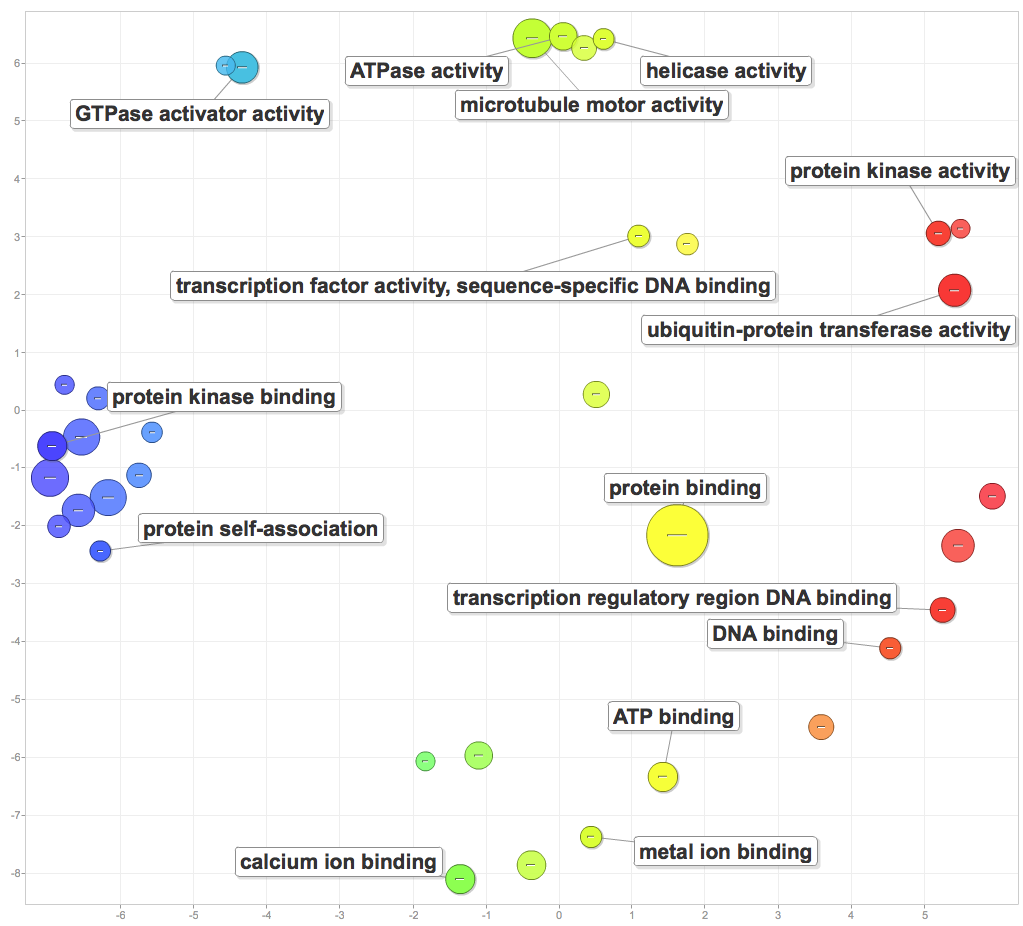
It’ll be interesting to see how the DAVID gene enrichment compares to the Mike Riffle’s gene enrichment tool, or how gene enrichment in the other components will complicate the story. For now, I have enough to work with for PCSGA.