DML Analysis Part 9
Gene Enrichment Results from compGO
Mike fixed the gene enrichment tool, so I got to use it! Good thing, becuase ReVIGO is currently down and I cannot generate gene enrichment visualizations any other way. All of this information can be found in my R Markdown analysis notebook.
| The website expects inputs in the sp | Q9C098 | DCLK3_HUMAN format. |

Figure 1. compGO interface.
This means I don’t need to unfold the specific code from anything. The tool generates both reports and images, all of which can be found in this folder. I can also do comparisons between two different lists, which is useful. I used a p-value of 1e-2, the default, for each enrichment.
DML-mRNA overlaps
Overrepresented biological processes include cytoskeletal growth and reorganization, transcription and translation regulation, and metabolic processes. I had no terms relating to methylation in this enrichment, but I did in my DAVID enrichment. This is probably due to the differences in p-value cutoffs I used with each method.
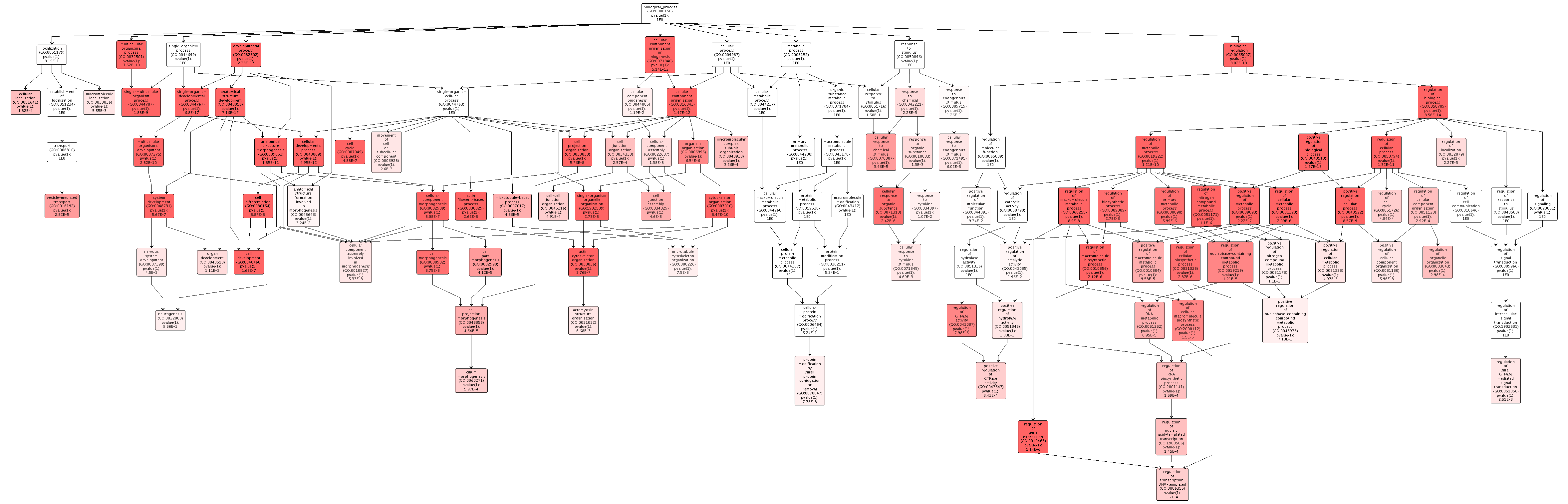
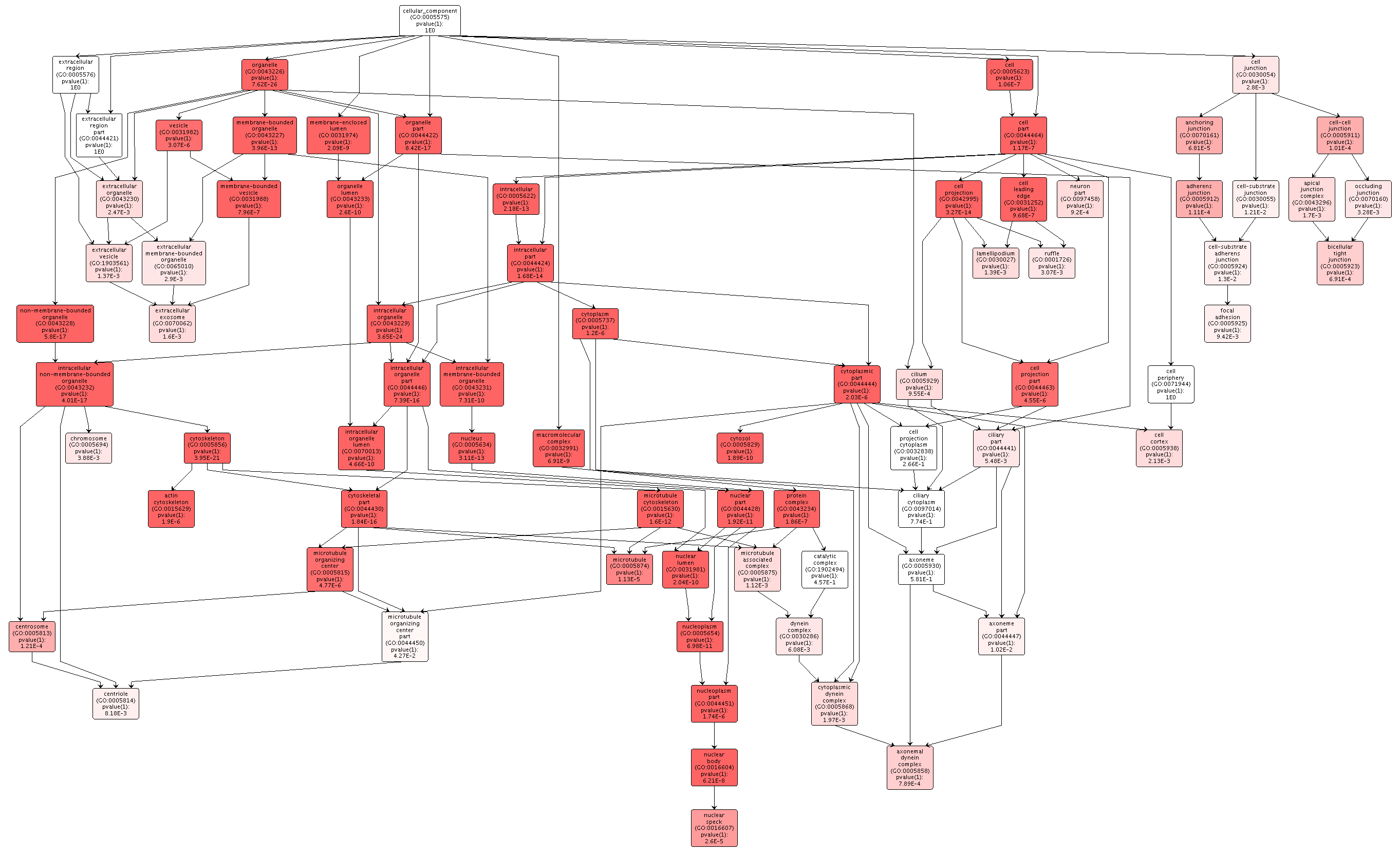
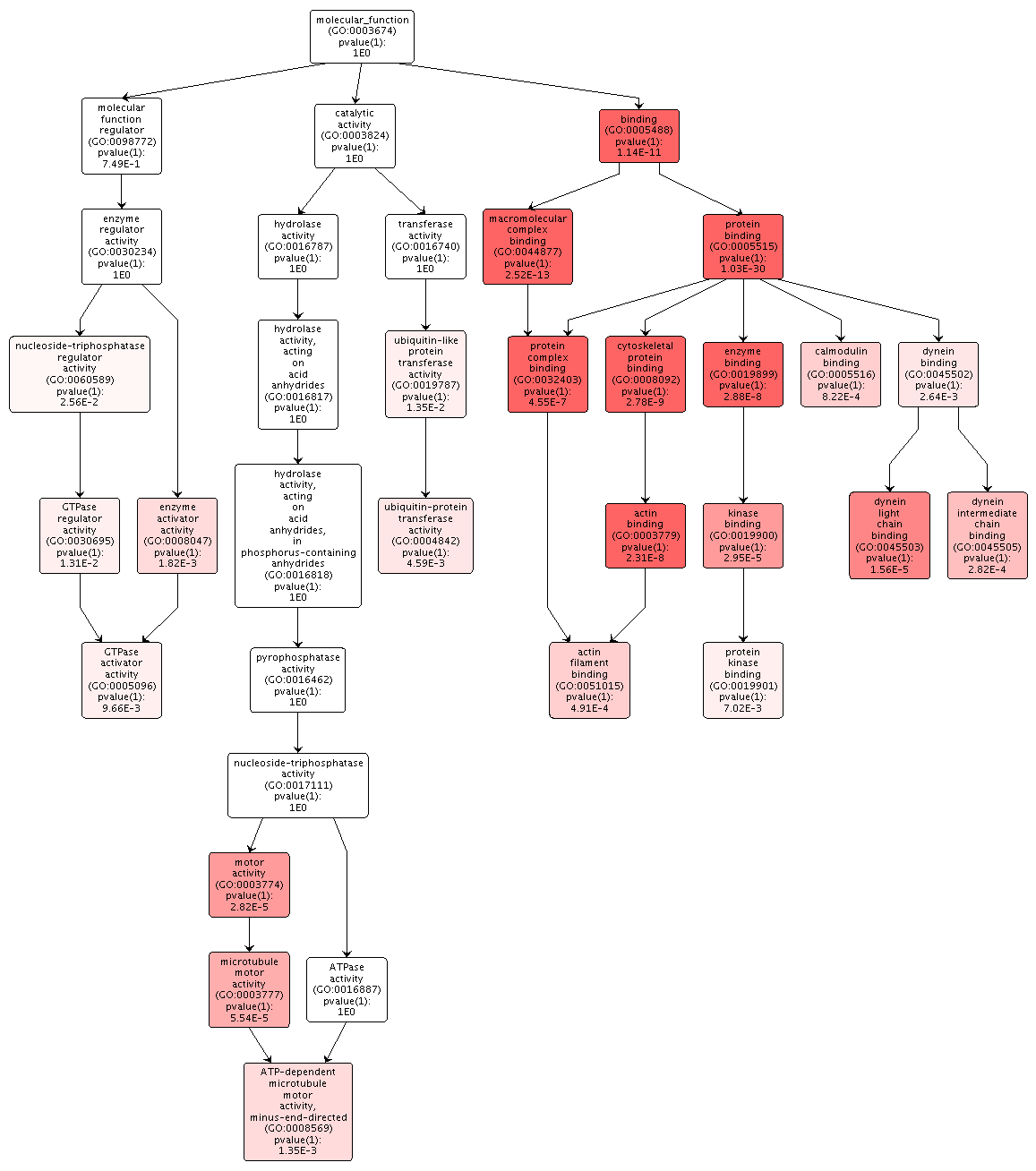
Figures 2-4. Gene enrichment visualizations for biological processes, cellular components, and molecular functions.
Exon and intron Uniprot codes
To get Uniprot codes for DML-Exon and DML-Intron overlaps, I used intersectBed with the -wao argument to find overlaps between these two files and the mRNA gff. -wao allowed me to write out the overlaps between the DML-Exon or DML-Intron file with the mRNA and count the number of overlaps. If there was no overlap (which was unlikely), then the DML-Exon or DML-Intron file information was written out with NULL to indicate that particular exon or intron had no overlap in the mRNA file.
I tried using the Uniprot code list from the DML-Exon file, but it kept crashing the tool. I found that the DML-Exon file is really large. There seem to be a bunch of repeating lines. Not sure if that’s just because several genome chunks lead to different mRNAs, and thus, different exon patterns, or if there was a problem with the -wao argument. If it’s just a problem with file size, I’ll need to ask Mike if that’s something he can accomodate. This is something I will need to investigate more after PCSGA.
DML-Intron overlaps
The overrepresented biological processes in the DML-Intron overlaps were focused on cellular components, like microtubules, organization and regulation of those components, and developmental processes. There was no indication of transcription or translation processes.
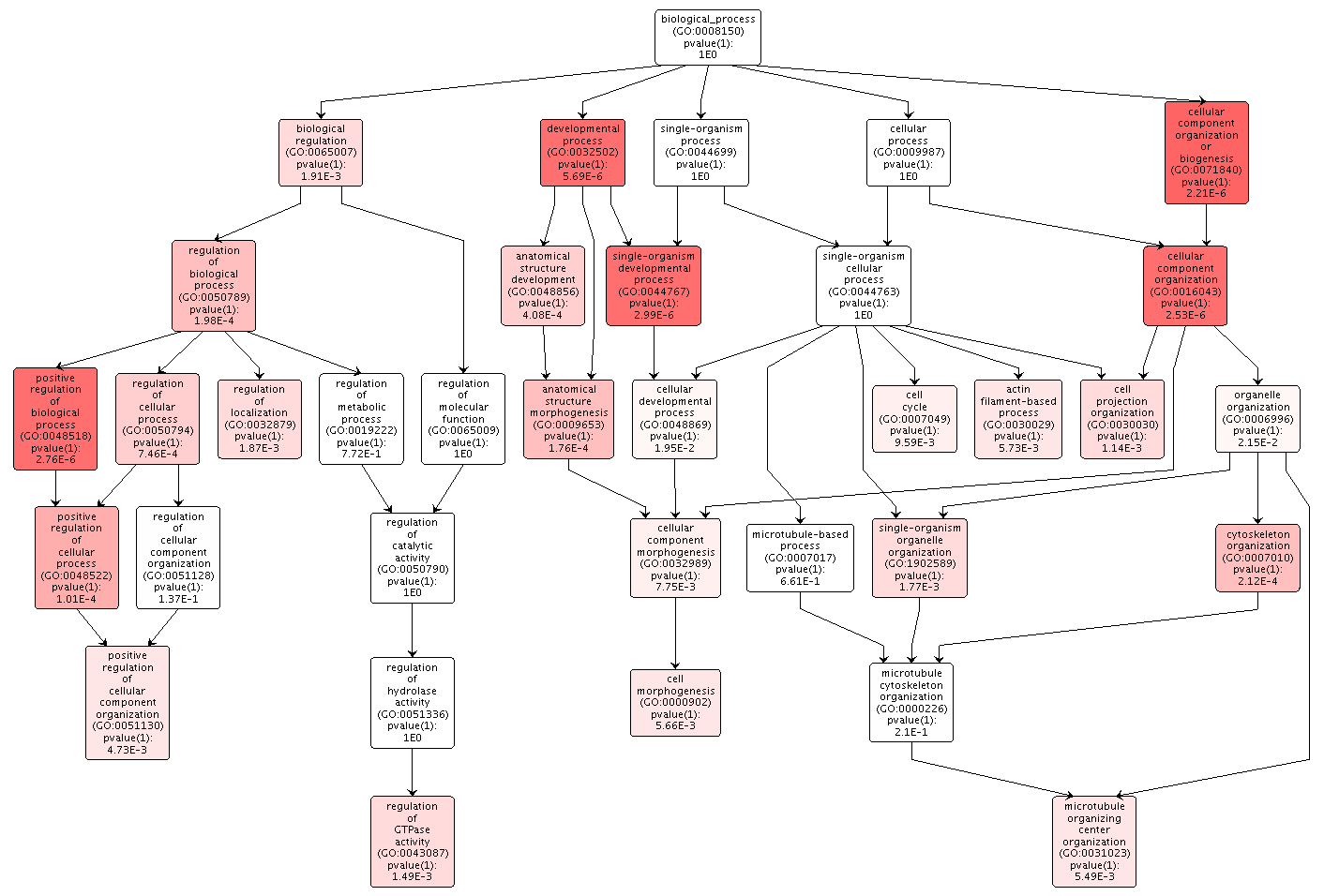
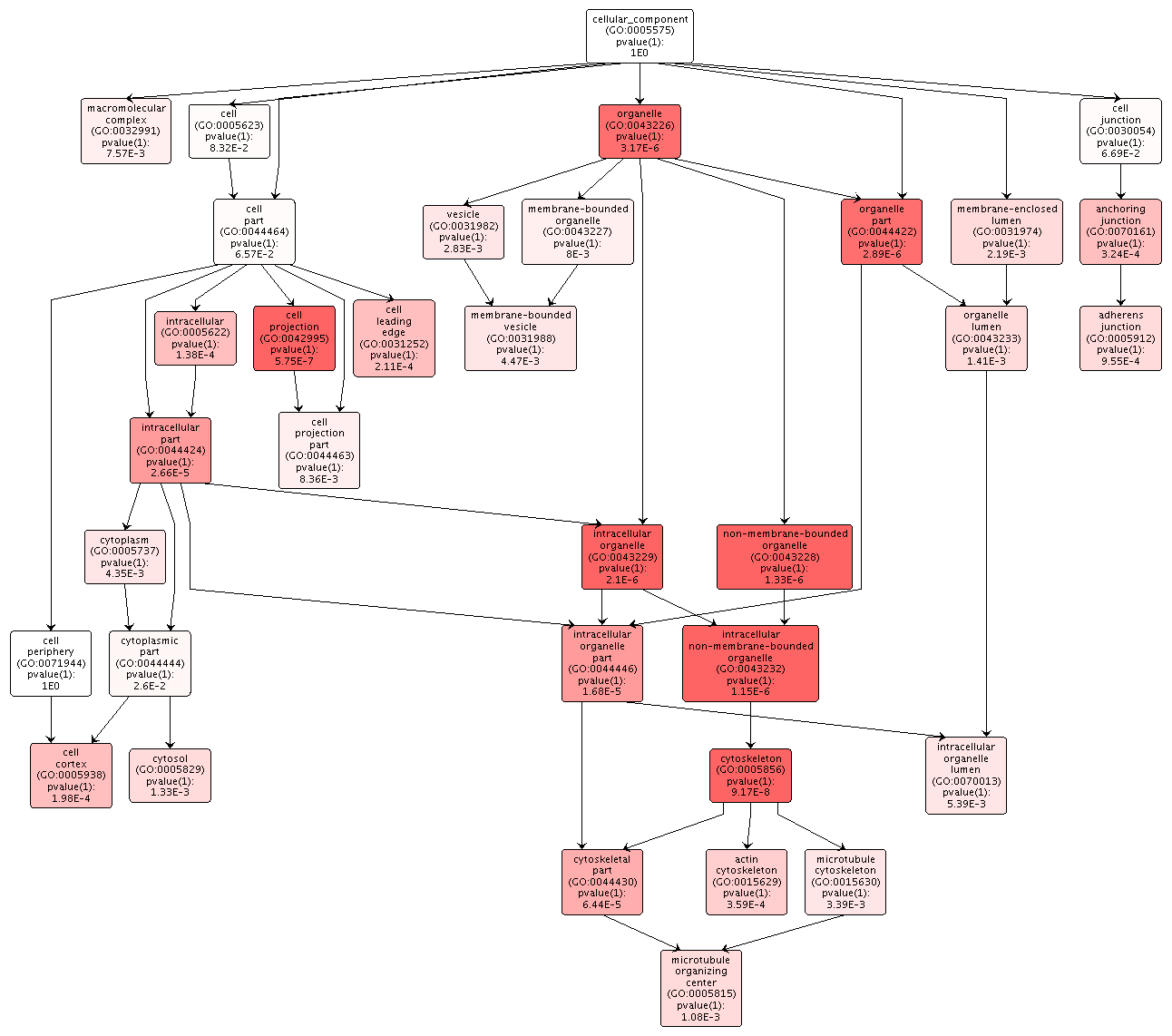
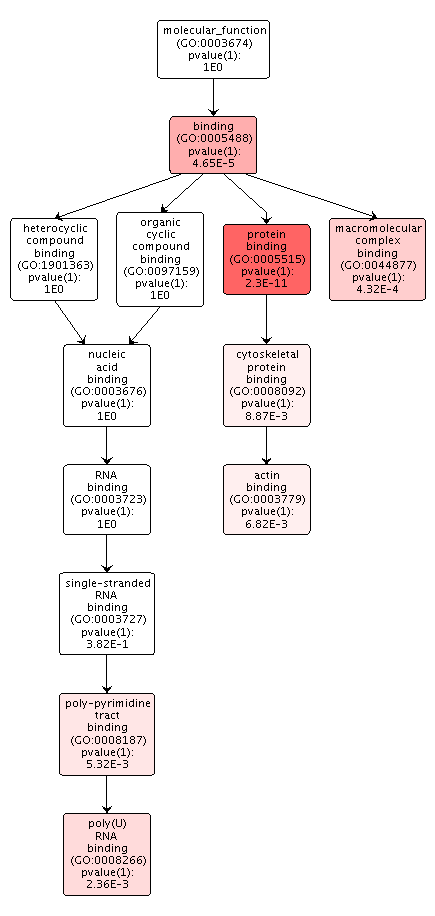
Figures 5-7. Gene enrichment visualizations for biological processes, cellular components, and molecular functions.
Biological processes comparison
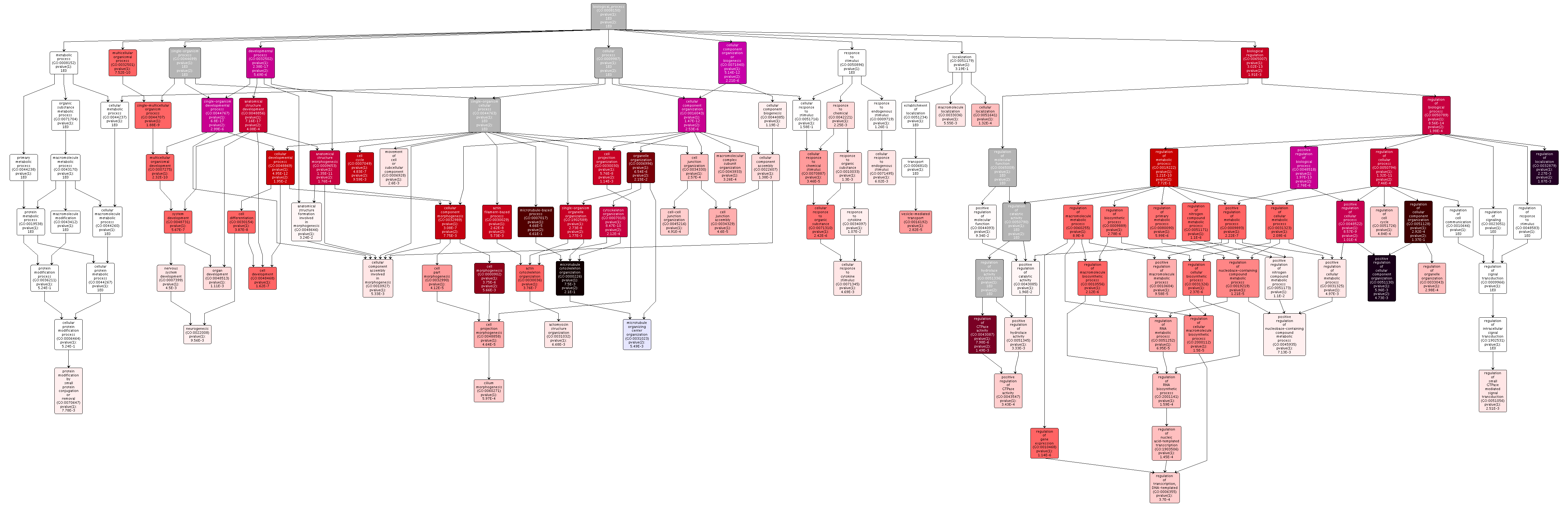
Figure 8. Comparison of DML-mRNA and DML-Intron gene enrichment.
Going forward
For PCSGA:
- Focus on
compGOoutput for DML-mRNA gene enrichment for main story
For the paper:
- Figure out problem with DML-Exon overlap and get
compGOoutput - Flanking element gene enrichment
- All component gene enrichment
- Compare all gene enrichment results