Gonad Methylation Analysis Part 18
Preparing for discoveries
Before I can understand where differentially methylated loci (DML) are located within the C. virginica genome, I first need to identify DMLs! I used this R script to identify DMLs that were at least 50% different between control and treatment (high pCO2) samples (.csv here). Looking at the PCA was interesting, as the clustering was not as tight as I expected.
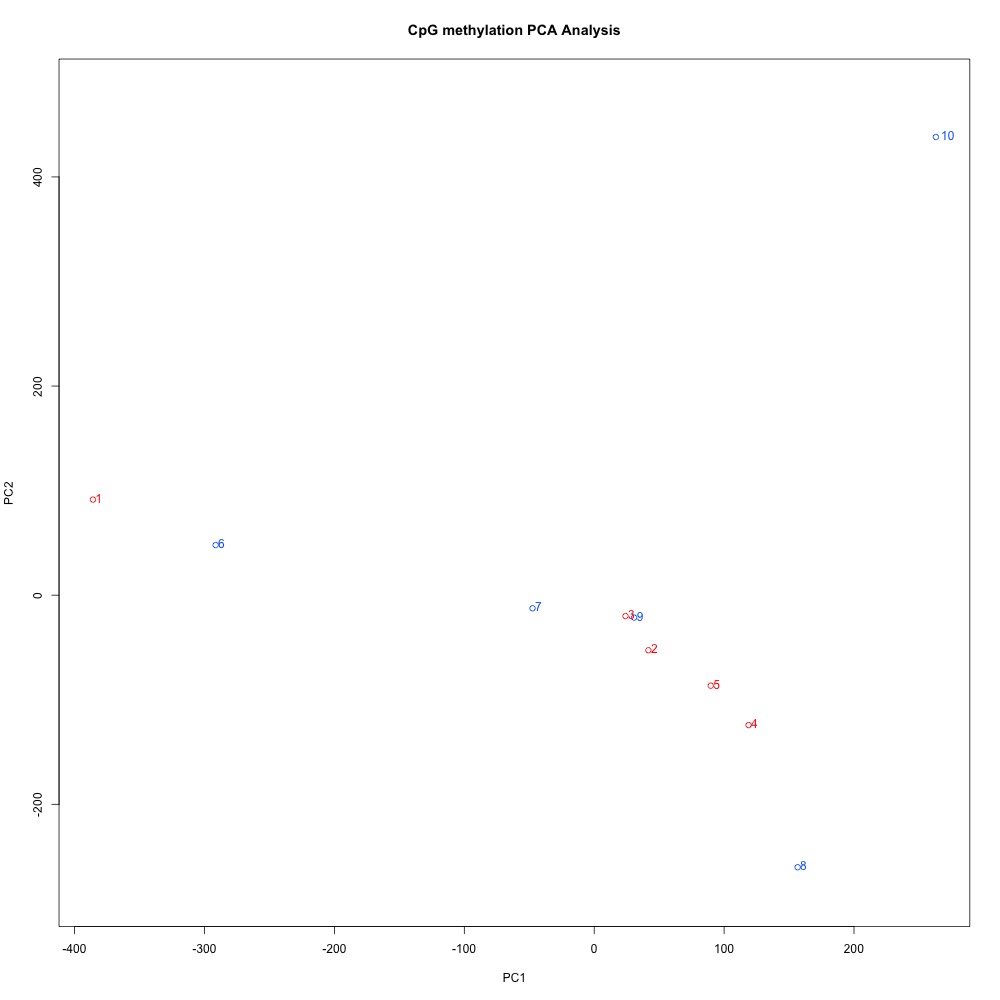
Figure 1. Principal Components Analysis of methylated regions in samples.
O2-5 (ambient conditions) are more closely clustered than any of the oysters from treatment conditions. It’s possible that there are organismal differences in methylation responses, or that we just didn’t have a large enough sample size to deal with this variation.
In the last part of the script, I saved my DML information as a BED file. I mimicked Steven’s code to do this. BED files have chromosome ID, start, and stop positions that I can use to pare down information about my DMLs. I can then compare the DML location with other important genomic features using bedtools. The intersect tool seems especially useful. I know we covered bedtools in the 2016 Bioinformatics class, so I’ll review those notes!