Remaining Analyses
Squeezing every last bit of information out of my data
As I outlined in my November Goals, this week I’m playing around with my dataset to see if I can pool my eelgrass and bare samples, and repeating my NMDS and ANOSIM analyses. I know Steven said to focus on protein-level responses, but I’m curious…
Step 1: Rerun NMDS and ANOSIM without pooling
Site only ANOSIM: R = 0.01339, Significance: 0.326 Eelgrass/Bare only ANOSIM: R = 0.02932, Significance: 0.168 Site and eelgrass ANOSIM: R = 0.00386, Significance: 0.41
Nothing significant, but here’s the pattern I see in that NMDS:
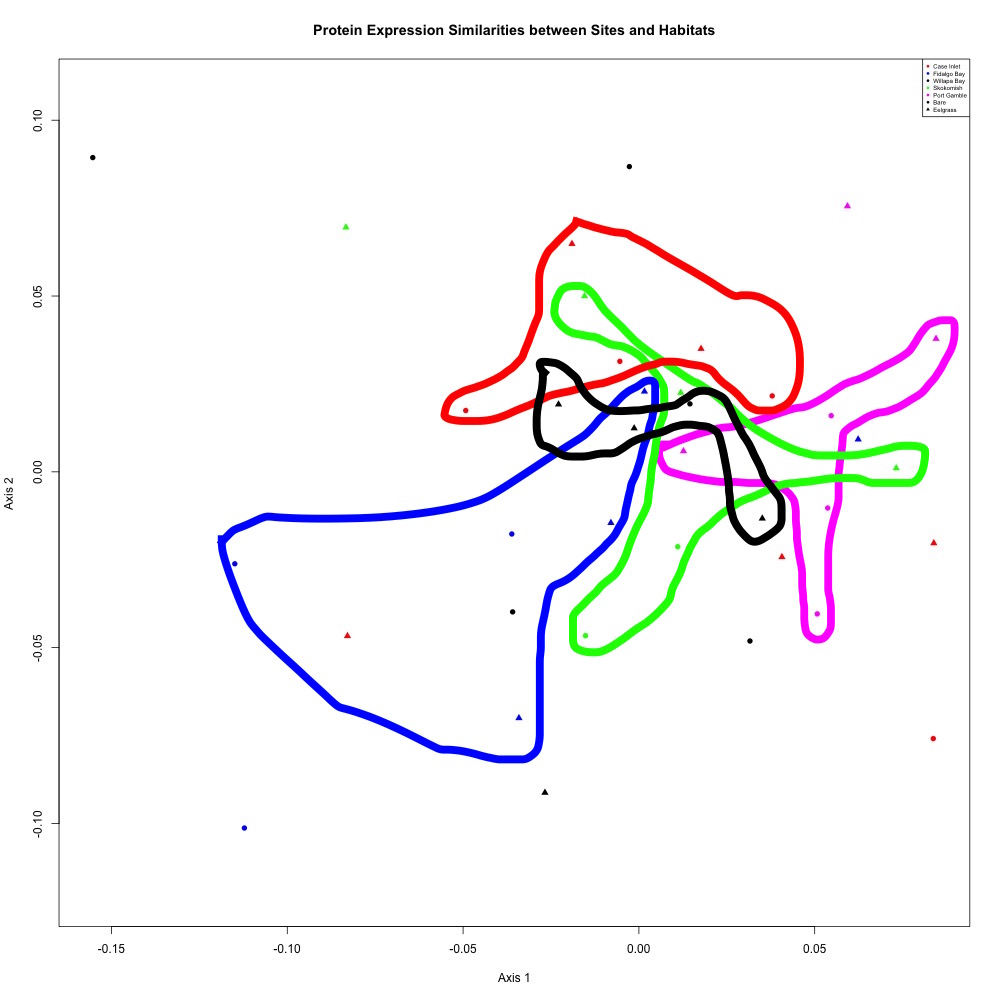
Figure 1. Annotated NMDS plot
My groups overlap because there’s still a lot of variability within my groups. This could be due to the alternative splicing I’m seeing within my protein abundances.
Step 2: Pool eelgrass and bare samples
There has been no indication that my eelgrass samples have significantly different protein expression results from my bare samples (see above eelgrass-only ANOSIM results). So I pooled them. Again nothing significant, but it’s nice looking at a less-distracting picture.
ANOSIM: R = 0.01339, Significance: 0.321
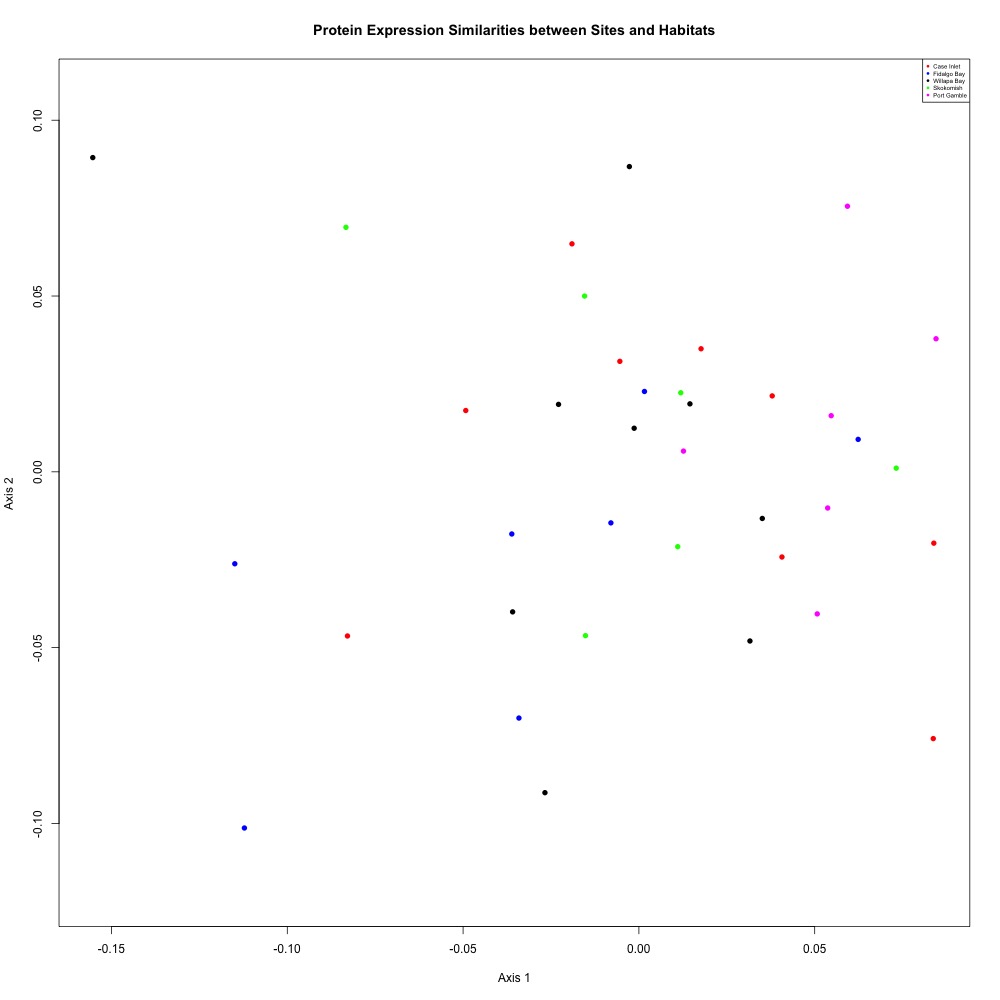
Figure 2. Annotated NMDS plot after pooling bare and eelgrass samples
I’m just go ahead and say we’re about done here. No more NMDS and ANOSIMs!