SRM Analysis Part 5
Everything sucks.
I normalized my data using TIC values, saved in my sequence file. After dividing all of my area values by TIC content, I remade my NMDS plot. Things did not look much better :disappointed:
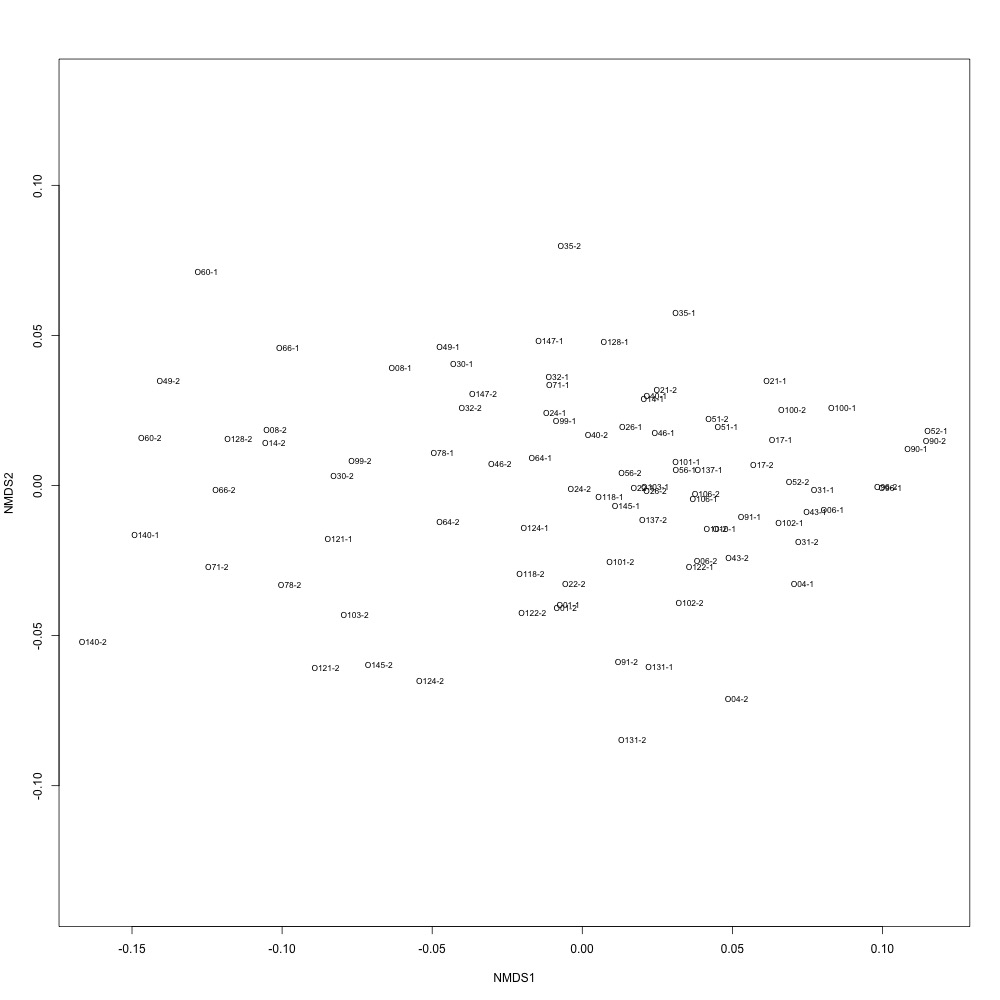
During my meeting with Julian, he suggested I calculate the distances between ordinations for each technical replicate so I have a quantitative way of pulling out bad technical replicate clustering. For my bad NMDS, the distances didn’t look great :cold_sweat:
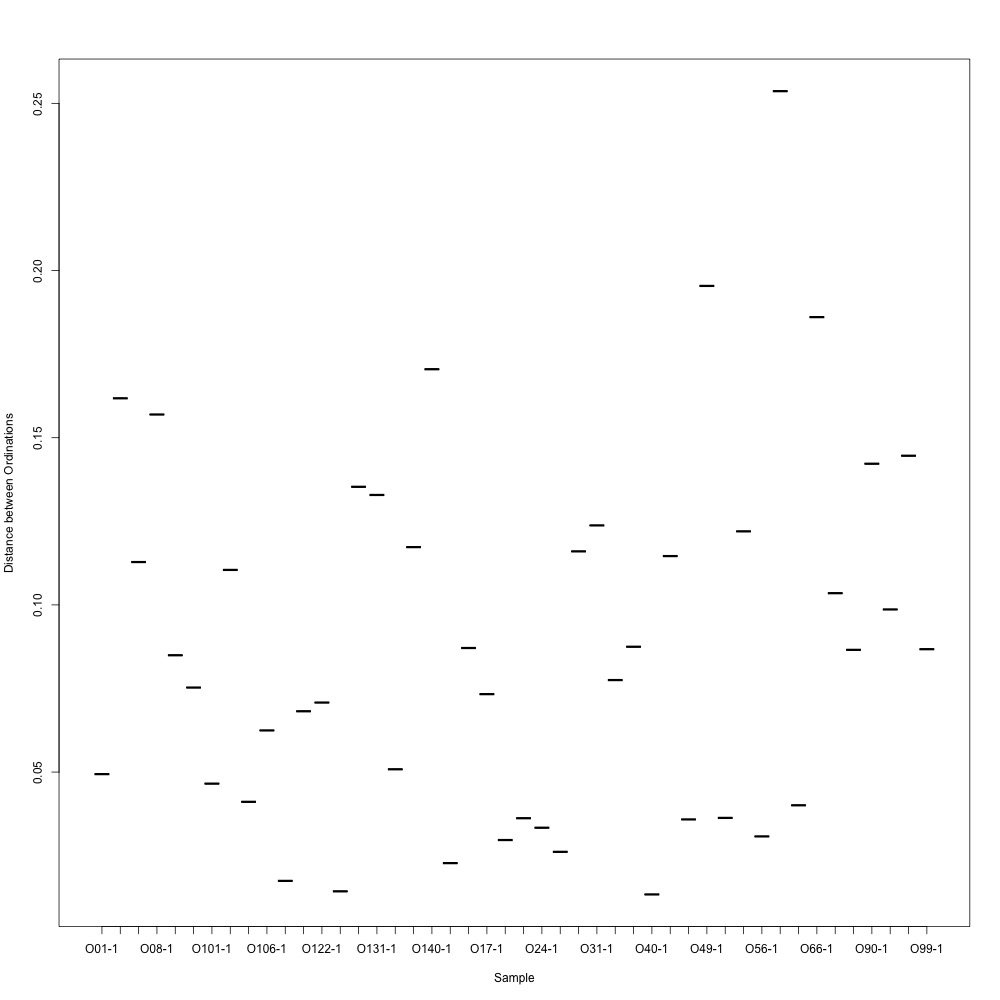
Emma said she’s never seen such bad technical replication. Will I have yet another failed experiment? Will I be able to present anything at PCSGA? Who could never be sure :sob:
Written on September 8, 2017