SRM Analysis Part 7
Another Skyline saga
To figure out what was going on with my technical replicates, Emma said she’d take a look at my Skyline document. Here’s what she found:
I found some discrepancies between your settings and settings I had for one of my SRM files (one of which was that you told Skyline you had DIA data…). I’ve updated the settings and I am re-importing your data. In peptide settings, the 13C modifications weren’t checked (PRTC modifications). In transition settings, mine looked like the attached, which were different from yours. I think the most significant would be the Full-scan settings.
I’m not sure why my Skyline settings were off! Emma and I made the Skyline document together, but we may not have actually adjusted the settings. Here are my old, incorrect Skyline settings:
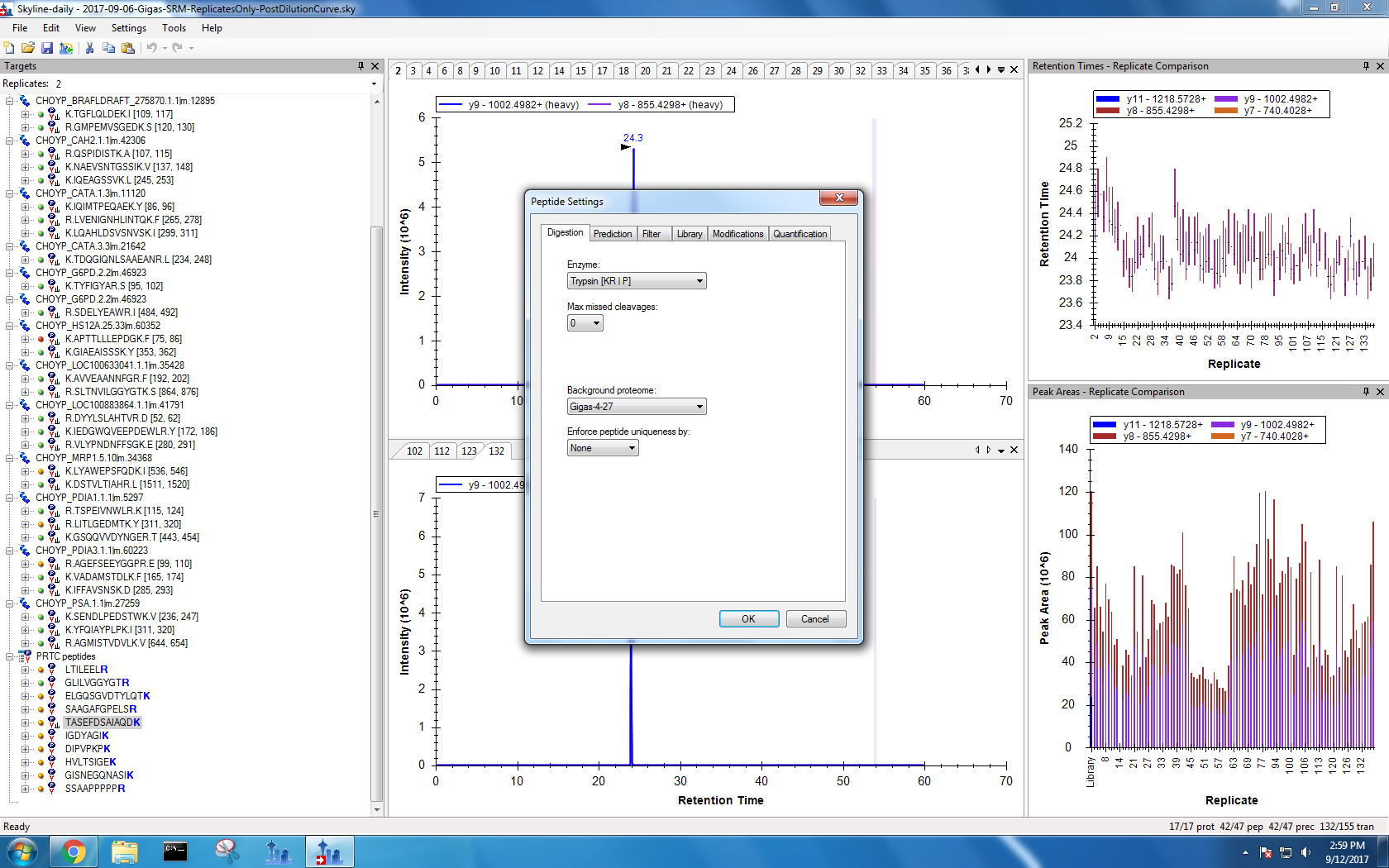







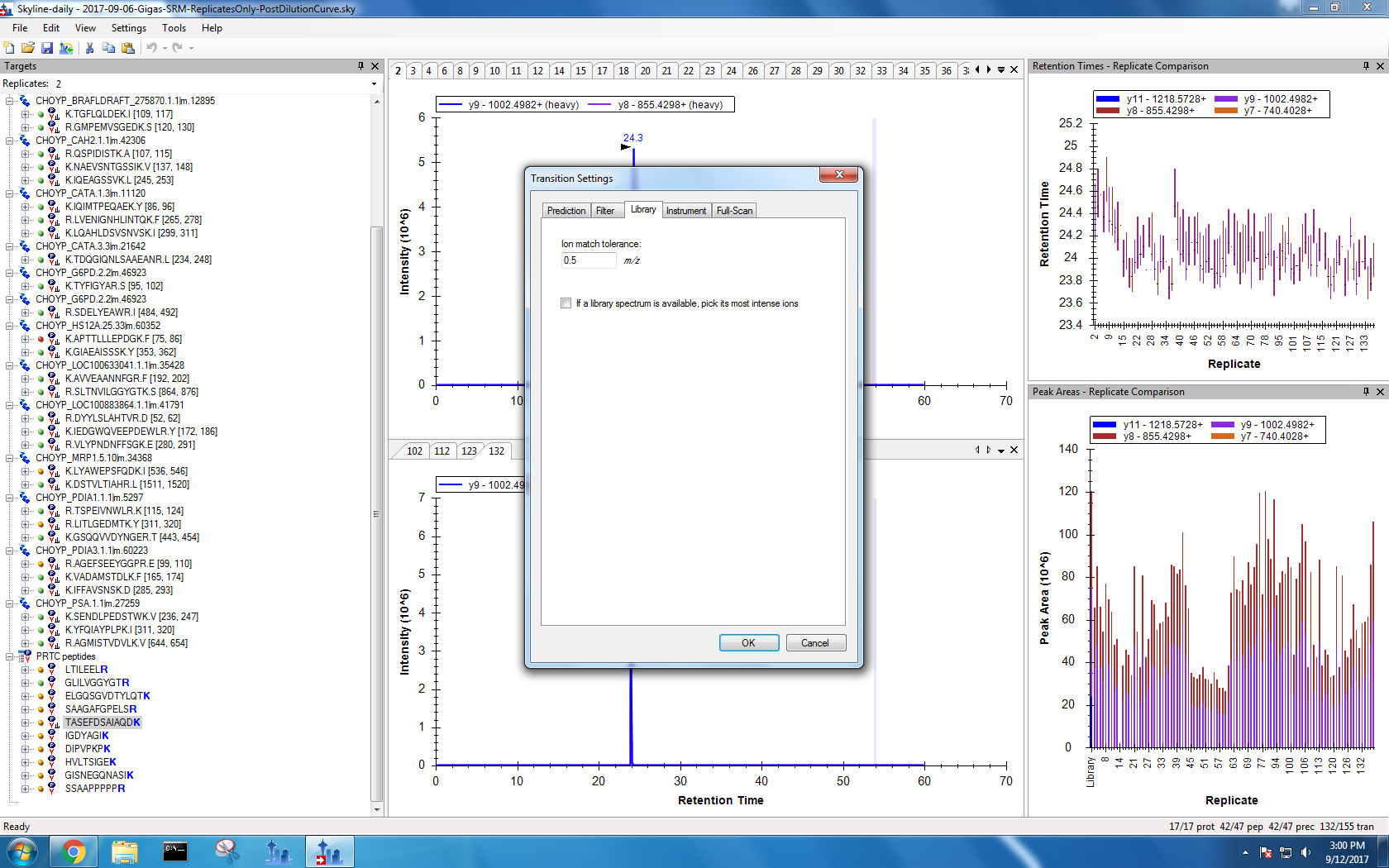


Figures 1-11. Old Skyline settings
And here are the actual settings:




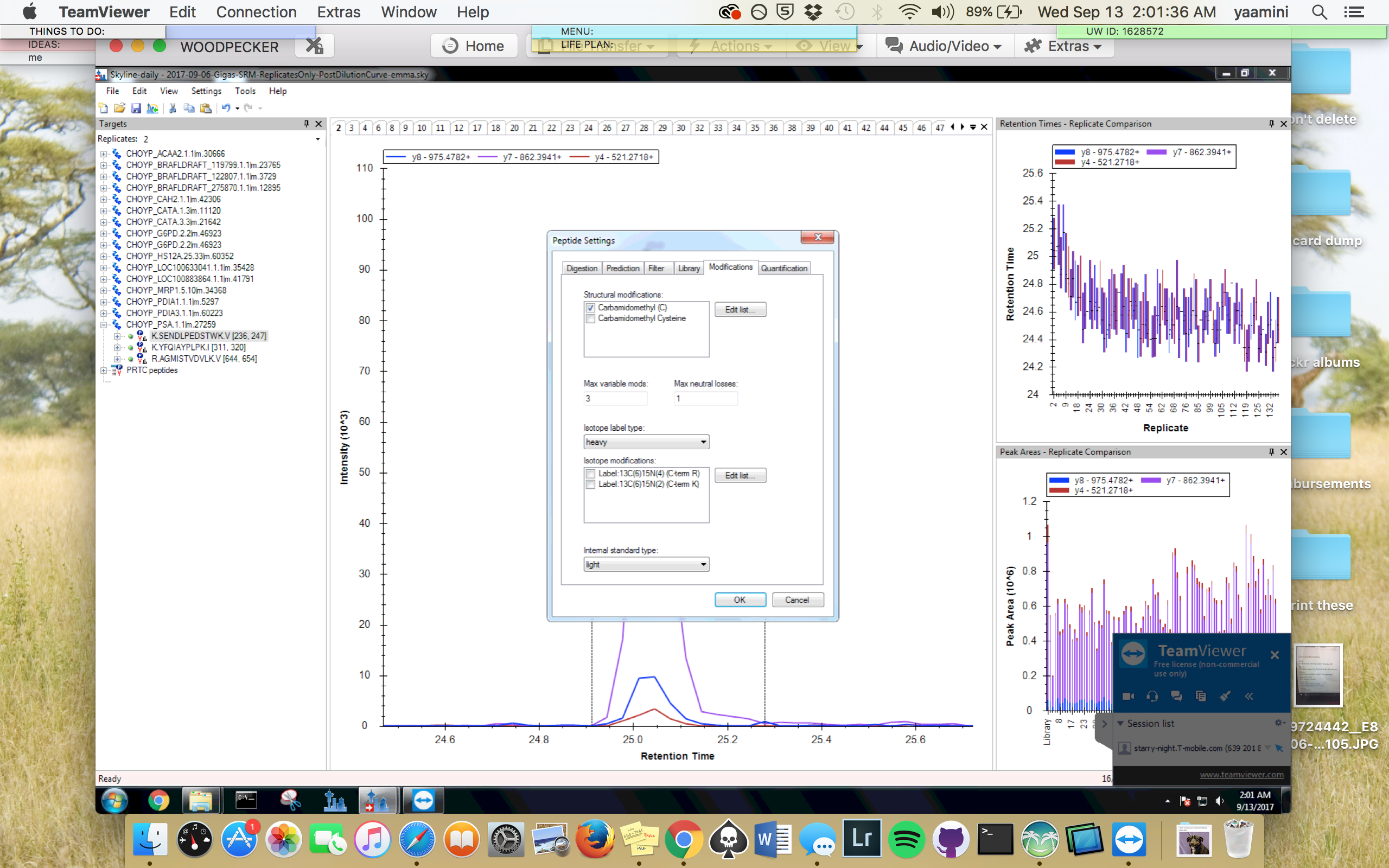






Figures 12-22. Revised Skyline settings
Once again, I fixed all transitions and ensured all peaks were picked correctly using my old Skyline document. Emma was unable to find any chromatograms in samples 14 and 15.




Figures 23-26. Examples of transitions that needed to be fixed.
After I cleaned the data, I uploaded my Skyline document to OWL and exported my area data.
I ran through my NMDS and ANOSIM scripts again to see if anything changed. The new settings slightly improved clustering, but not enough for me to feel confident in my technical replication.
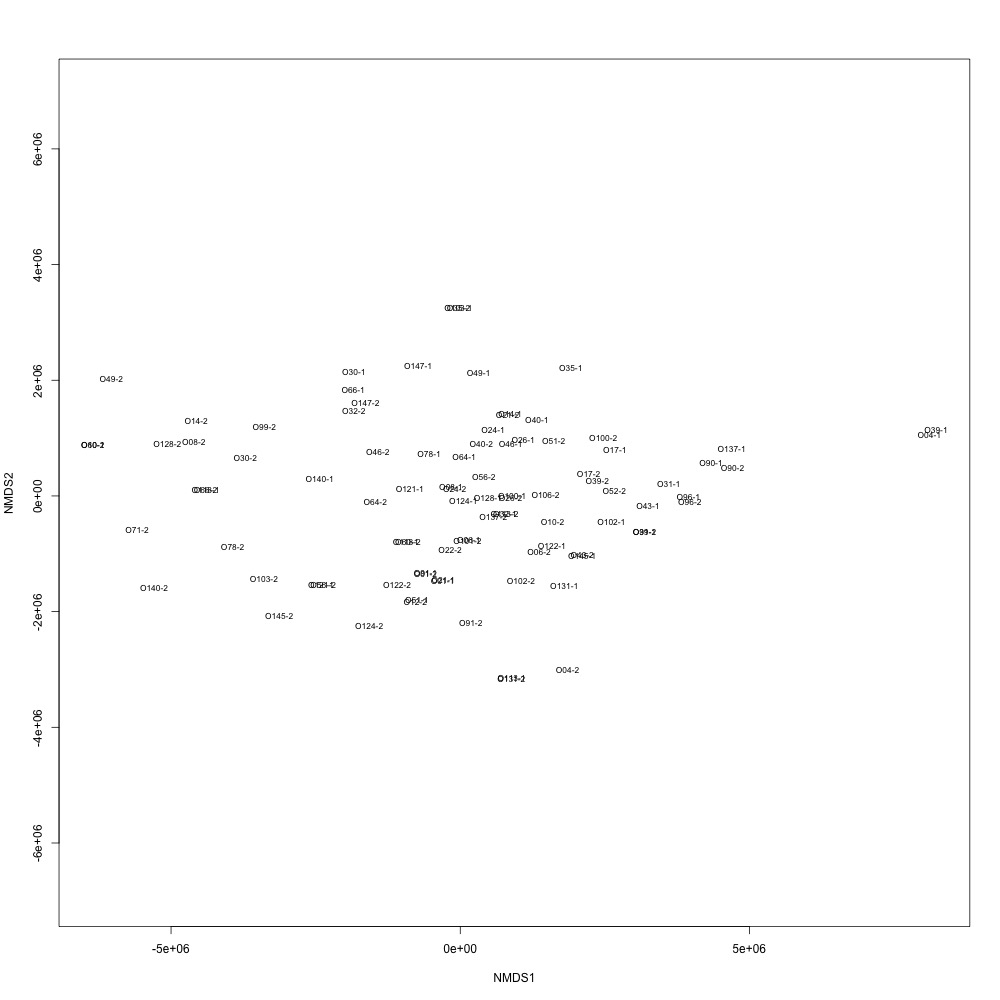
Figure 27. NMDS for technical replication using nonnormalized data.
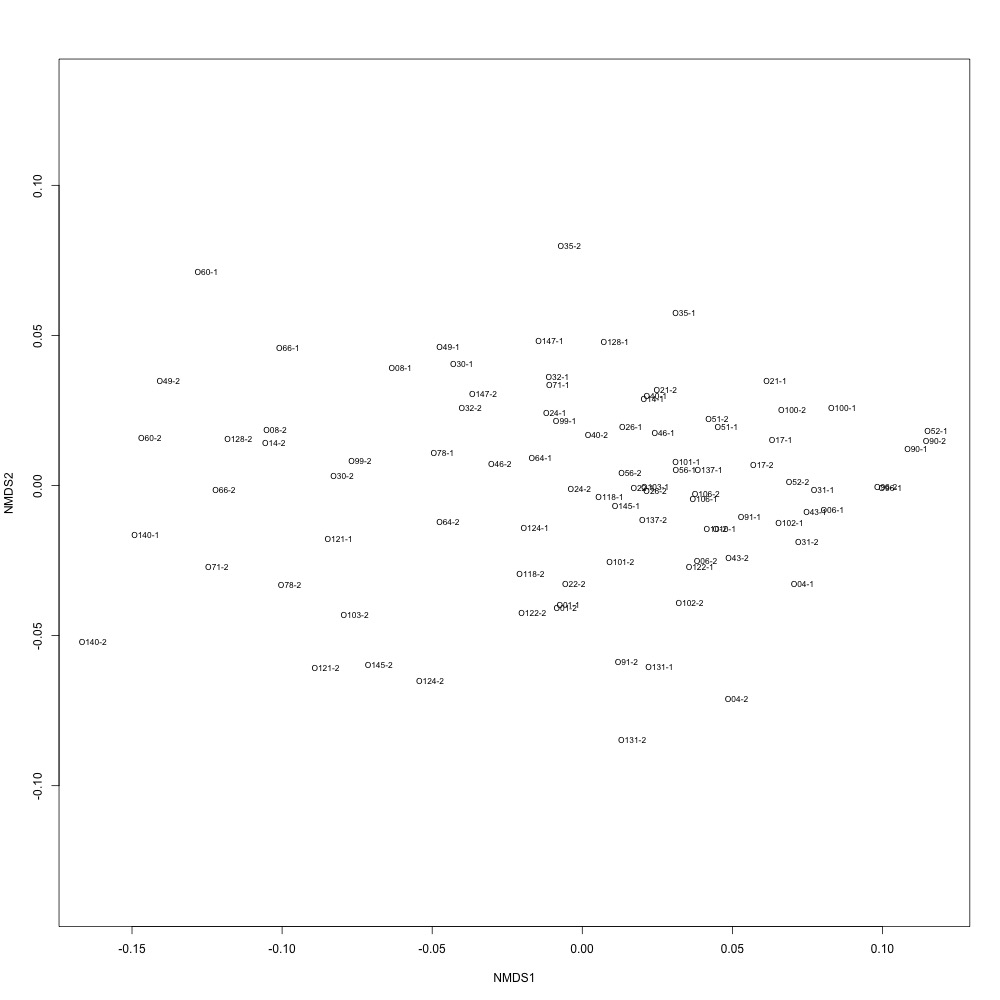
Figure 28. NMDS for technical replication using normalized data.
Now I just need to figure out how to analyze and present my SRM data for PCSGA…………