Skyline Attempt 3
Remember that Skyline fail?
We think it’s fixed! Based on my previous error rates, Emma and her team were able to figure out that brecan (a version of pecanpie produced by Brian Searle in Genome Sciences) “produces blib files that have a certain number of incompatibilities with Skyline (one of which is redundant peptide identifications). Jarrett Egertson wrote a script to get rid of these incompatibilities so that the brecan blib files work,” (Timmins-Schiffman, pers. comm.). I saved the .blib file here in OWL.
Using Skyline document settings from my test document and first full Skyline attempt, and results specifications from my first try at SRM target selection, I created a new Skyline document.
Step 1: Peptide settings
I used the proteome file from my Gigas-4-27 document:




Added the new “desearlinated” .blib:

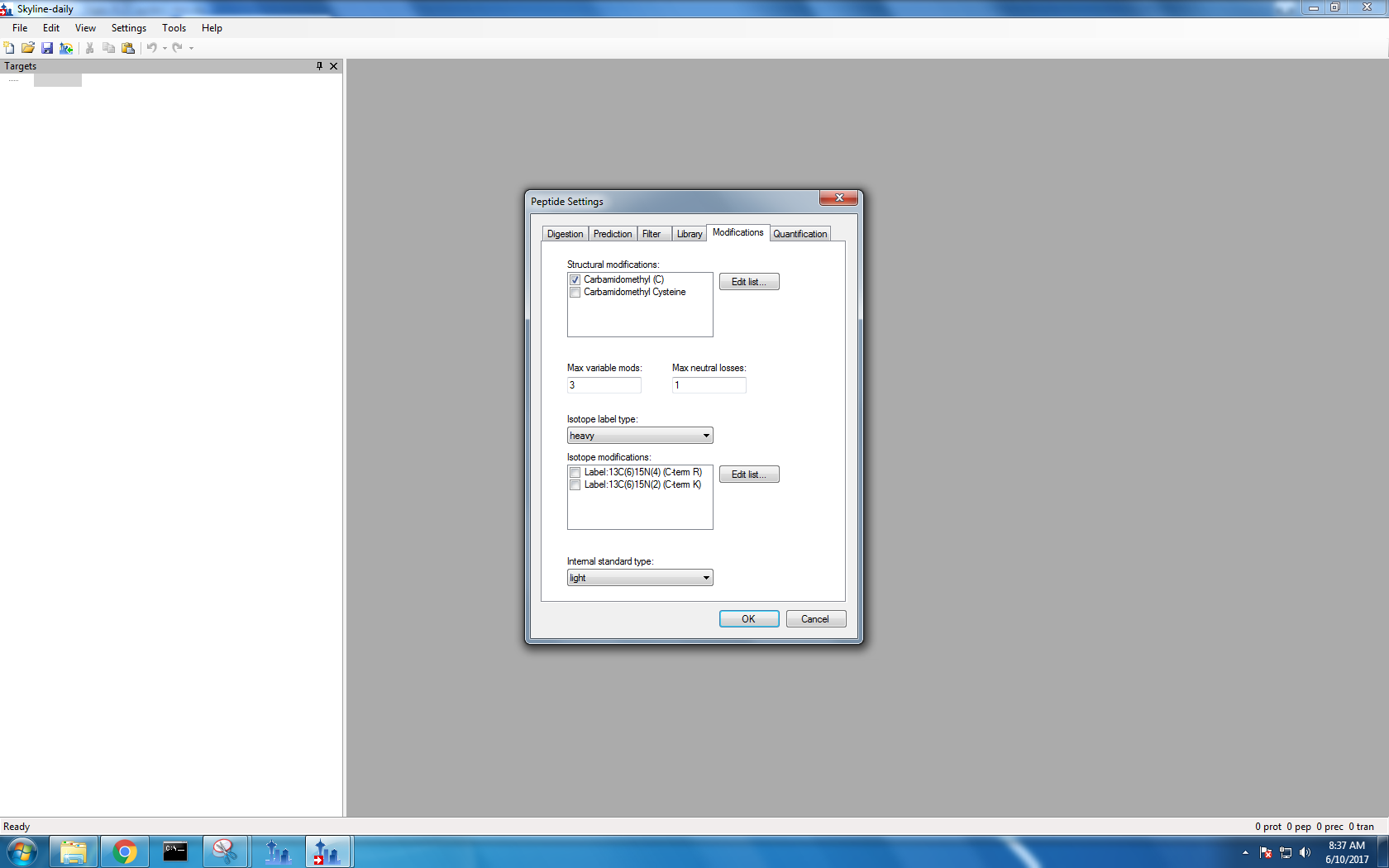

Step 2: Import FASTA file
File » Import » FASTA » Combined-gigas-QC.txt
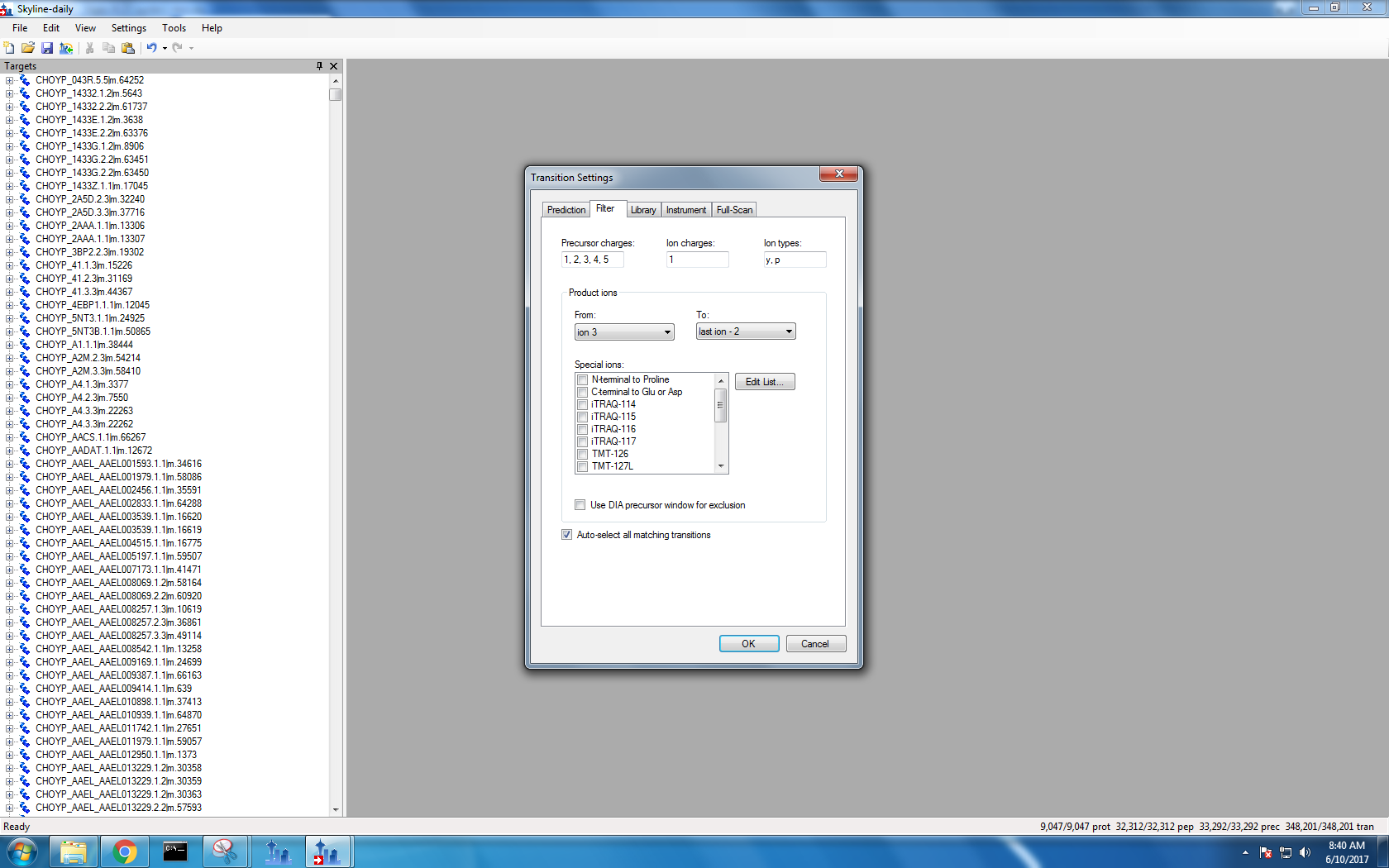
Step 3: Transition settings




Step 4: Import demultiplexed .mzmL files
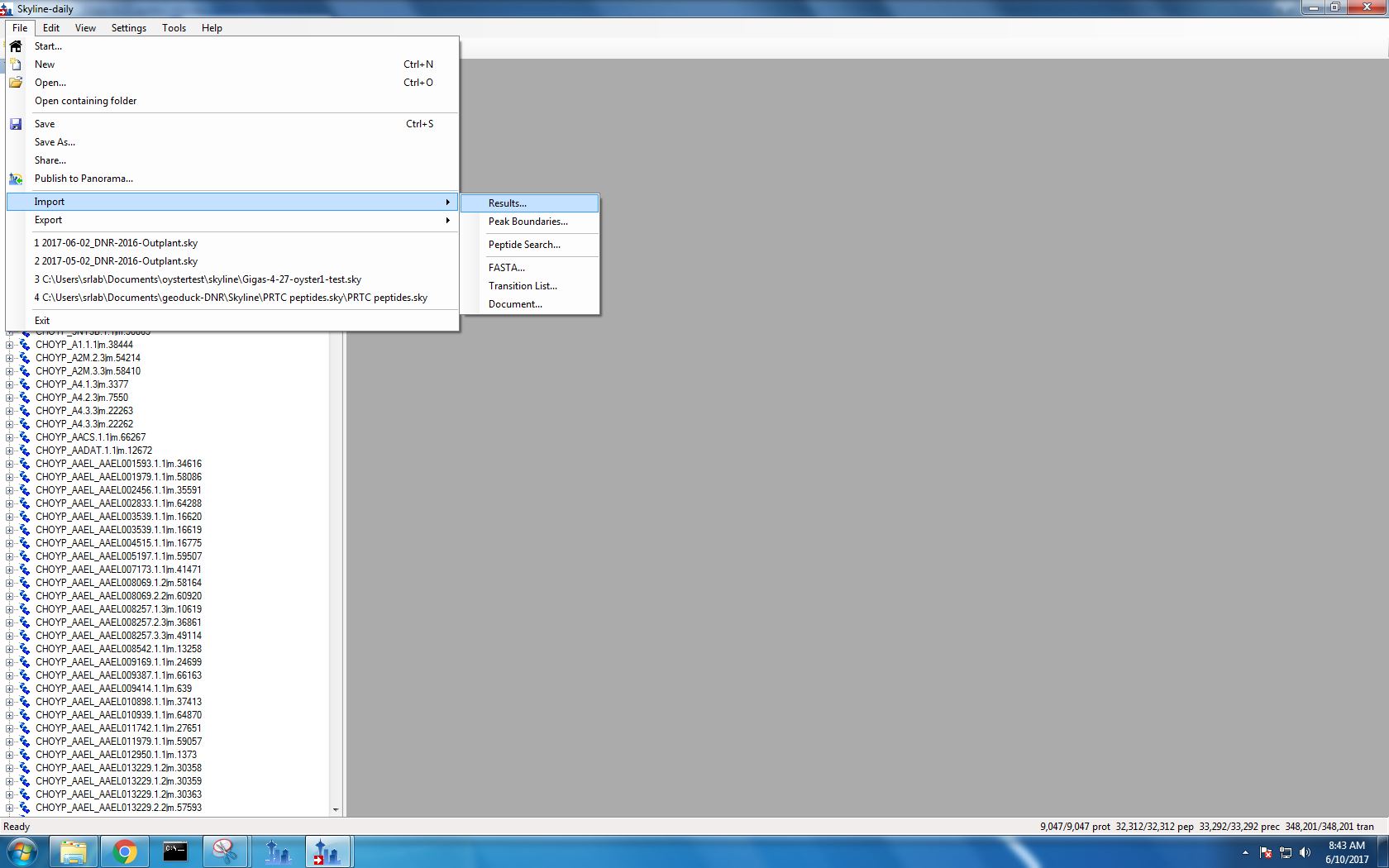
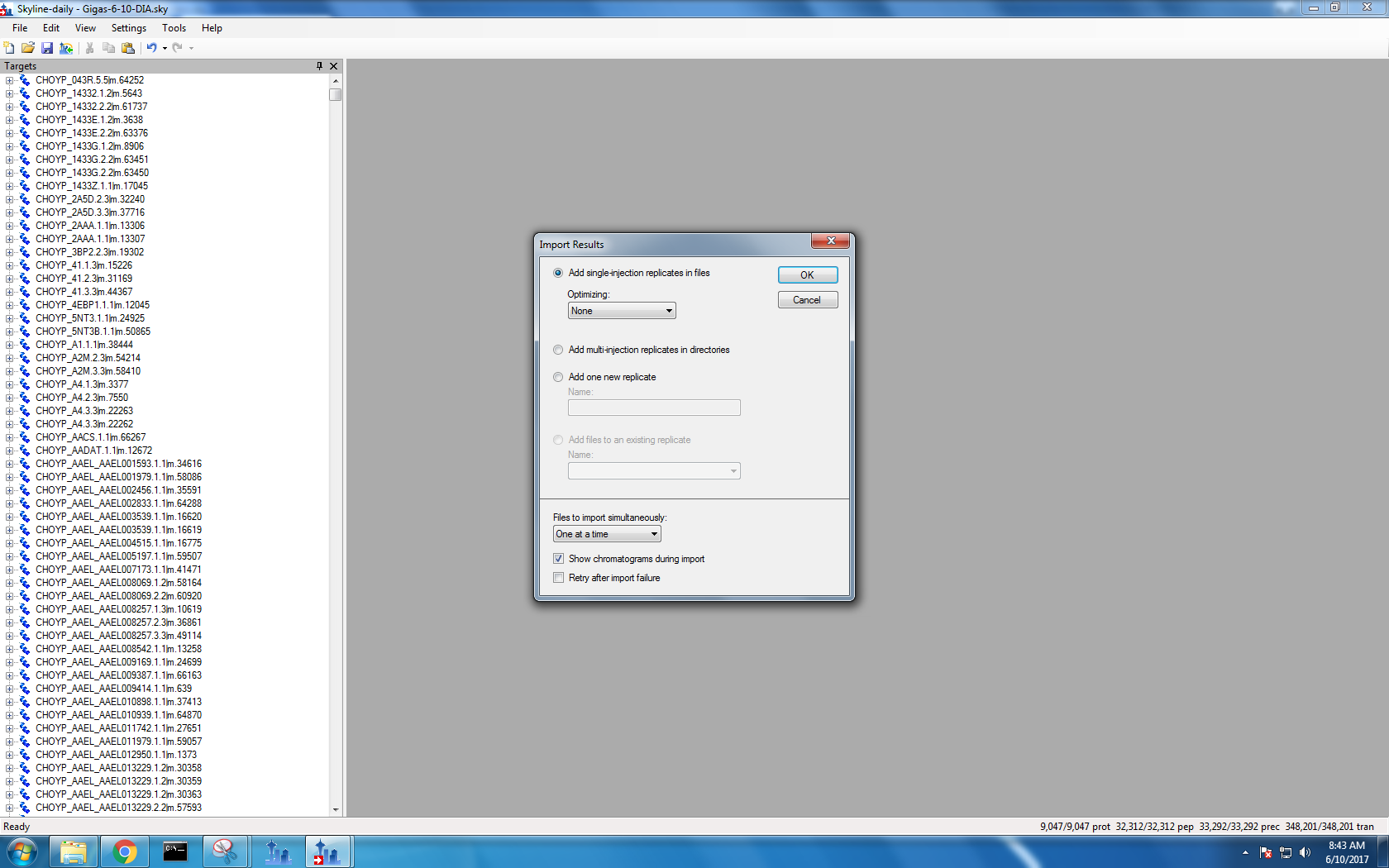

I didn’t upload files 17, 21, 22 and 23 because oyster 21 was my blank (so there should be no data in it!) and oyster 17, 22 and 23 corresponded with my original 0107 sample had a bubble and did not read properly in the mass spec.



Once my data files finish loading, I’m going to start error checking again to see how much of a difference the new .blib made.
Step 5: Add Condition and BioReplicate information
Based on information from this lab notebook.



Step 6: Export Skyline report three separate ways

Just proteins and peak areas, with replicate pivoted

File found on OWL here
For MSstats
Edit “Transition Results” » “Transition Results”


Replicate pivoted:

File found here. This was actually a mistake, but I saved it anyways.
Replicate not pivoted:

File found here. This is what I can use for MSstats.
Now I’m ready to error check.