Virginica MBDSeq
All systems go on C. Virginica sequencing!
Today Sam and I visited Mac at NOAA Montlake to shear DNA in preparation for MBDSeq lab work next week. Here’s an overview of the entire process:
Step 1: Isolate and quantify DNA. Sam did this previously.
- Isolate DNA - Use E.Z.N.A. Kit (Omega)
- Quantify DNA - Use Qubit dsDNA Kit (Invitrogen/ThermoFisher)
Step 2: Shear DNA to about 500 bp. Not all DNA with have methylated cytosines. By cutting the DNA into smaller fragments, we can wash away DNA that doesn’t have any methylated cytosines and enrich the methylation signal. This is what we did today!
- Shear DNA to target fragment length (~500bp) - Use NOAA sonicator
- Verify shearing - Use Seeb Lab Bioanlyzer with DNA 12000 Kit (Agilent)
Step 3: Methylation enrichment. We’re doing this next week.
- Enrich for MBD - Use MethylMiner Kit (Invitrogen/ThermoFisher)
- Quantify DNA - Use Qubit hsDNA Kit (Invitrogen/ThermoFisher)
Back to what we did today.
We loaded eight samples into the QSONICA CD0004054245, which consists of a sample holder and a water bath. The sonicator blasts sound waves at our samples for ten minutes, alternating between 30 second on and off periods at 25% intensity and 4ºC. However, with our first eight samples, the machine spit out an “overload” error.
Figure 1. Overload error from sonicator.
Mac looked at the manual, and apparently there are tons of reasons why this could happen. They range from machine parts not being tight enough, to having too much water or electrical issues. We couldn’t figure out what was wrong. More importantly, we didn’t know how many cycles our samples went through. Pushing our samples through an additional ten cycles would make our DNA too small. We restarted the machine and ran through four more cycles. After the fourth “on” phase, the machine overloaded again. We assume it did something similar when we first turned it on.
Just to test the machine, we loaded eight blanks with 350 µL of water in the sonicator. We ran through five cycles, and the machine did not produce an overload error. Maybe the difference in sample volume (350 µL blank vs. 100 µL of DNA) could have produced the issue?
We then loaded our last two samples, 106 and 108 into the machine. We ran through five cycles before the machine produced the same overload error. We then restarted the machine and ran through five more cycles, stopping the machine after the fifth off cycle.
To verify shearing (and see if our first eight samples were sheared to the proper length), we used the Agilent Technologies 2200 TapeStation. Mac pipetted our samples into a different set of sample wells and mixed them with the necessary reagents. However, I distracted her while she was pipetting, so she accidentally pipetted two samples into the same well. Luckily, these samples were from the first round of unknown sonication! So she wasn’t mixing samples we know were sheared properly with those that weren’t.

Table 1. TapeStation sample set-up.
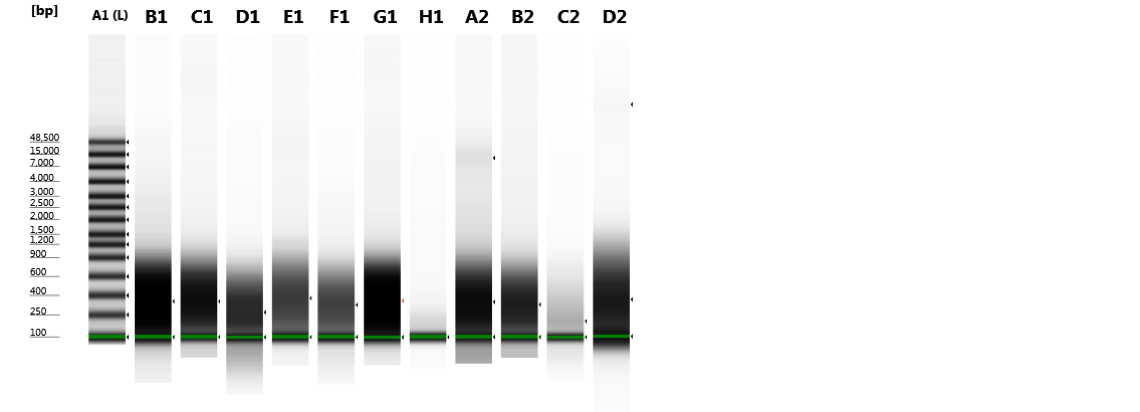
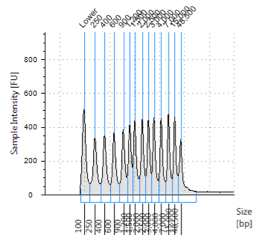

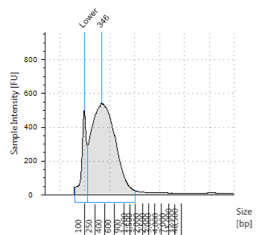
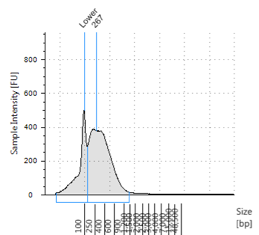
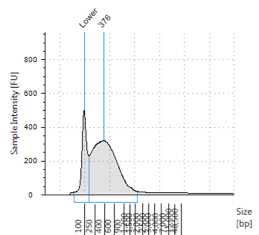
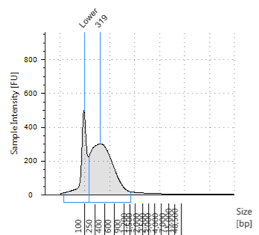
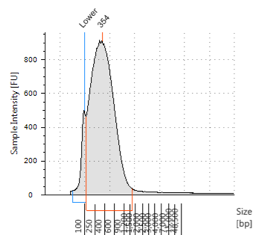
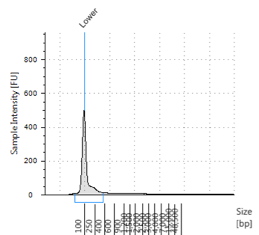
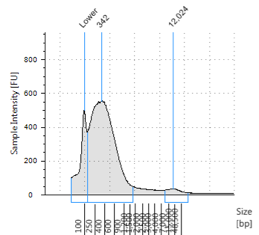
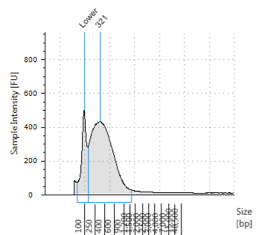
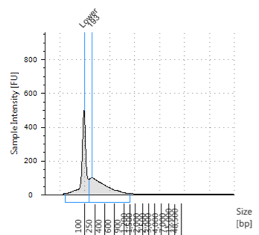
Figures 2-13. Shearing results. The peak indicates the average DNA fragment length. A1 was the ladder, which we used to estimate the length of sample DNA. G1 had two samples while H1 was blank because I distracted Mac while she was pipetting.
Looking at our results (word document and .gdna file), we saw that most of our samples had average lengths around 350 bp. Sam deemed this good enough for us! Next week, we’ll do the actual methylation enrichment.